Role of the transcription activator Ste12p as a repressor of PRY3 expression
- PMID: 16940175
- PMCID: PMC1636733
- DOI: 10.1128/MCB.01004-06
Role of the transcription activator Ste12p as a repressor of PRY3 expression
Abstract
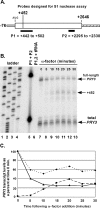
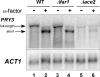
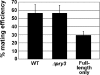
Mating pheromone represses synthesis of full-length PRY3 mRNA, and a new transcript appears simultaneously with its 5' terminus 452 nucleotides inside the open reading frame (ORF). Synthesis of this shorter transcript results from activation of a promoter within the PRY3 locus, and its production is concomitant with the rapid disappearance of the full-length transcript. Evidence is consistent with the pheromone-induced transcription factor Ste12p binding two pheromone response elements within the PRY3 promoter, directly impeding transcription of the full-length mRNA while simultaneously inducing initiation of the short transcript. This process depends on a TATA box within the PRY3 ORF. Expression of full-length PRY3 inhibited mating, while no disadvantage was detectable for cells unable to make the short transcript. Therefore, Ste12p is utilized as a repressor of full-length PRY3 transcription, ensuring efficient mating. There is no evidence that production of the short PRY3 transcript is anything more than an adventitious by-product of this mechanism. It is possible that cryptic binding sites for transcriptional activators may occur frequently within genomes and have the potential of evolving for rapid, gene-specific repression by mechanisms analogous to PRY3. PRY3 regulation provides a model for the coordination of both inductive and repressive activities within a regulatory network.
Figures


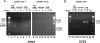
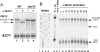

Similar articles
-
Transcriptional activation upon pheromone stimulation mediated by a small domain of Saccharomyces cerevisiae Ste12p.Mol Cell Biol. 1997 Nov;17(11):6410-8. doi: 10.1128/MCB.17.11.6410. Mol Cell Biol. 1997. PMID: 9343403 Free PMC article.
-
The Zap1 transcriptional activator negatively regulates translation of the RTC4 mRNA through the use of alternative 5' transcript leaders.Mol Microbiol. 2017 Dec;106(5):673-677. doi: 10.1111/mmi.13856. Epub 2017 Oct 26. Mol Microbiol. 2017. PMID: 28971534 Free PMC article.
-
Zap1-dependent transcription from an alternative upstream promoter controls translation of RTC4 mRNA in zinc-deficient Saccharomyces cerevisiae.Mol Microbiol. 2017 Dec;106(5):678-689. doi: 10.1111/mmi.13851. Epub 2017 Oct 23. Mol Microbiol. 2017. PMID: 28963784 Free PMC article.
-
Signaling activation and repression of RNA polymerase II transcription in yeast.Bioessays. 1997 Nov;19(11):1001-10. doi: 10.1002/bies.950191110. Bioessays. 1997. PMID: 9394622 Review.
-
Functionality of unspliced XBP1 is required to explain evolution of overlapping reading frames.Trends Genet. 2006 Dec;22(12):645-8. doi: 10.1016/j.tig.2006.09.012. Epub 2006 Oct 10. Trends Genet. 2006. PMID: 17034899 Review.
Cited by
-
The yeast cell wall protein Pry3 inhibits mating through highly conserved residues within the CAP domain.Biol Open. 2020 Jul 2;9(6):bio053470. doi: 10.1242/bio.053470. Biol Open. 2020. PMID: 32554483 Free PMC article.
-
An evolutionarily 'young' lysine residue in histone H3 attenuates transcriptional output in Saccharomyces cerevisiae.Genes Dev. 2011 Jun 15;25(12):1306-19. doi: 10.1101/gad.2050311. Genes Dev. 2011. PMID: 21685365 Free PMC article.
-
Chromatin- and transcription-related factors repress transcription from within coding regions throughout the Saccharomyces cerevisiae genome.PLoS Biol. 2008 Nov 11;6(11):e277. doi: 10.1371/journal.pbio.0060277. PLoS Biol. 2008. PMID: 18998772 Free PMC article.
-
Intragenic transcription of a noncoding RNA modulates expression of ASP3 in budding yeast.RNA. 2010 Nov;16(11):2085-93. doi: 10.1261/rna.2177410. Epub 2010 Sep 3. RNA. 2010. PMID: 20817754 Free PMC article.
-
Control and signal processing by transcriptional interference.Mol Syst Biol. 2009;5:300. doi: 10.1038/msb.2009.61. Epub 2009 Aug 18. Mol Syst Biol. 2009. PMID: 19690569 Free PMC article.
References
-
- Adhya, S., and M. Gottesman. 1982. Promoter occlusion: transcription through a promoter may inhibit its activity. Cell 29:939-944. - PubMed
-
- Basehoar, A. D., S. J. Zanton, and B. F. Pugh. 2004. Identification and distinct regulation of yeast TATA box-containing genes. Cell 116:699-709. - PubMed
-
- Beltzer, J. P., S. R. Morris, and G. B. Kohlhaw. 1988. Yeast LEU4 encodes mitochondrial and nonmitochondrial forms of alpha-isopropylmalate synthase. J. Biol. Chem. 263:368-374. - PubMed
-
- Boguta, M., L. A. Hunter, W. C. Shen, E. C. Gillman, N. C. Martin, and A. K. Hopper. 1994. Subcellular locations of MOD5 proteins: mapping of sequences sufficient for targeting to mitochondria and demonstration that mitochondrial and nuclear isoforms commingle in the cytosol. Mol. Cell. Biol. 14:2298-2306. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
