Alpha2,3 and alpha2,6 N-linked sialic acids facilitate efficient binding and transduction by adeno-associated virus types 1 and 6
- PMID: 16940521
- PMCID: PMC1563919
- DOI: 10.1128/JVI.00895-06
Alpha2,3 and alpha2,6 N-linked sialic acids facilitate efficient binding and transduction by adeno-associated virus types 1 and 6
Abstract
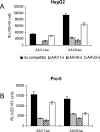
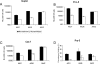
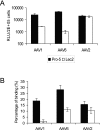
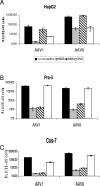
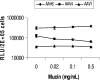
Recombinant adeno-associated viruses (AAVs) are promising vectors in the field of gene therapy. Different AAV serotypes display distinct tissue tropism, believed to be related to the distribution of their receptors on target cells. Of the 11 well-characterized AAV serotypes, heparan sulfate proteoglycan and sialic acid have been suggested to be the attachment receptors for AAV type 2 and types 4 and 5, respectively. In this report, we identify the receptor for the two closely related serotypes, AAV1 and AAV6. First, we demonstrate using coinfection experiments and luciferase reporter analysis that AAV1 and AAV6 compete for similar receptors. Unlike heparin sulfate, enzymatic or genetic removal of sialic acid markedly reduced AAV1 and AAV6 binding and transduction. Further analysis using lectin staining and lectin competition assays identified that AAV1 and AAV6 use either alpha2,3-linked or alpha2,6-linked sialic acid when transducing numerous cell types (HepG2, Pro-5, and Cos-7). Treatment of cells with proteinase K but not glycolipid inhibitor reduced AAV1 and AAV6 infection, supporting the hypothesis that the sialic acid that facilitates infection is associated with glycoproteins rather than glycolipids. In addition, we determined by inhibitor (N-benzyl GalNAc)- and cell line-specific (Lec-1) studies that AAV1 and AAV6 require N-linked and not O-linked sialic acid. Furthermore, a resialylation experiment on a deficient Lec-2 cell line confirmed a 2,3 and 2,6 N-linked sialic acid requirement, while studies of mucin with O-linked sialic acid showed no inhibition effect for AAV1 and AAV6 transduction on Cos-7 cells. Finally, using a glycan array binding assay we determined that AAV1 efficiently binds to NeuAcalpha2-3GalNAcbeta1-4GlcNAc, as well as two glycoproteins with alpha2,3 and alpha2,6 N-linked sialic acids. Taken together, competition, genetic, inhibitor, enzymatic reconstitution, and glycan array experiments support alpha2,3 and alpha2,6 sialic acids that are present on N-linked glycoproteins as primary receptors for efficient AAV1 and AAV6 viral infection.
Figures

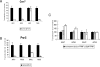



References
-
- Bantel-Schaal, U., and H. zur Hausen. 1984. Characterization of the DNA of a defective human parvovirus isolated from a genital site. Virology 134:52-63. - PubMed
-
- Blankinship, M. J., P. Gregorevic, J. M. Allen, S. Q. Harper, H. Harper, C. L. Halbert, D. A. Miller, and J. S. Chamberlain. 2004. Efficient transduction of skeletal muscle using vectors based on adeno-associated virus serotype 6. Mol. Ther. 10:671-678. - PubMed
-
- Blixt, O., S. Head, T. Mondala, C. Scanlan, M. E. Huflejt, R. Alvarez, M. C. Bryan, F. Fazio, D. Calarese, J. Stevens, N. Razi, D. J. Stevens, J. J. Skehel, I. van Die, D. R. Burton, I. A. Wilson, R. Cummings, N. Bovin, C. H. Wong, and J. C. Paulson. 2004. Printed covalent glycan array for ligand profiling of diverse glycan binding proteins. Proc. Natl. Acad. Sci. USA 101:17033-17038. - PMC - PubMed
-
- Bonsdorff, L. V., H. Tolo, E. Lindeberg, T. Nyman, A. Harju, and J. Parkkinen. 2001. Development of a pharmaceutical apotransferrin product for iron binding therapy. Biologicals 29:27-37. - PubMed
-
- Chao, H., Y. Liu, J. Rabinowitz, C. Li, R. J. Samulski, and C. E. Walsh. 2000. Several log increase in therapeutic transgene delivery by distinct adeno-associated viral serotype vectors. Mol. Ther. 2:619-623. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- GM 059299/GM/NIGMS NIH HHS/United States
- P01 HL051818/HL/NHLBI NIH HHS/United States
- HL 51811/HL/NHLBI NIH HHS/United States
- P50 HL059412/HL/NHLBI NIH HHS/United States
- K08 HL069973/HL/NHLBI NIH HHS/United States
- HL 069973/HL/NHLBI NIH HHS/United States
- HL 59412/HL/NHLBI NIH HHS/United States
- P01 GM059299/GM/NIGMS NIH HHS/United States
- P01 HL059412/HL/NHLBI NIH HHS/United States
- U54 GM062116/GM/NIGMS NIH HHS/United States
- P01 HL051811/HL/NHLBI NIH HHS/United States
- GM 62116/GM/NIGMS NIH HHS/United States
- HL 051818/HL/NHLBI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources

