Genome of horsepox virus
- PMID: 16940536
- PMCID: PMC1563943
- DOI: 10.1128/JVI.00945-06
Genome of horsepox virus
Abstract
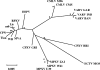
Here we present the genomic sequence of horsepox virus (HSPV) isolate MNR-76, an orthopoxvirus (OPV) isolated in 1976 from diseased Mongolian horses. The 212-kbp genome contained 7.5-kbp inverted terminal repeats and lacked extensive terminal tandem repetition. HSPV contained 236 open reading frames (ORFs) with similarity to those in other OPVs, with those in the central 100-kbp region most conserved relative to other OPVs. Phylogenetic analysis of the conserved region indicated that HSPV is closely related to sequenced isolates of vaccinia virus (VACV) and rabbitpox virus, clearly grouping together these VACV-like viruses. Fifty-four HSPV ORFs likely represented fragments of 25 orthologous OPV genes, including in the central region the only known fragmented form of an OPV ribonucleotide reductase large subunit gene. In terminal genomic regions, HSPV lacked full-length homologues of genes variably fragmented in other VACV-like viruses but was unique in fragmentation of the homologue of VACV strain Copenhagen B6R, a gene intact in other known VACV-like viruses. Notably, HSPV contained in terminal genomic regions 17 kbp of OPV-like sequence absent in known VACV-like viruses, including fragments of genes intact in other OPVs and approximately 1.4 kb of sequence present only in cowpox virus (CPXV). HSPV also contained seven full-length genes fragmented or missing in other VACV-like viruses, including intact homologues of the CPXV strain GRI-90 D2L/I4R CrmB and D13L CD30-like tumor necrosis factor receptors, D3L/I3R and C1L ankyrin repeat proteins, B19R kelch-like protein, D7L BTB/POZ domain protein, and B22R variola virus B22R-like protein. These results indicated that HSPV contains unique genomic features likely contributing to a unique virulence/host range phenotype. They also indicated that while closely related to known VACV-like viruses, HSPV contains additional, potentially ancestral sequences absent in other VACV-like viruses.
Figures


References
-
- Afonso, C. L., E. R. Tulman, Z. Lu, L. Zsak, N. T. Sandybaev, U. Z. Kerembekova, V. L. Zaitsev, G. F. Kutish, and D. L. Rock. 2002. The genome of camelpox virus. Virology 295:1-9. - PubMed
-
- Albagli, O., P. Dhordain, C. Deweindt, G. Lecocq, and D. Leprince. 1995. The BTB/POZ domain: a new protein-protein interaction motif common to DNA- and actin-binding proteins. Cell Growth Differ. 6:1193-1198. - PubMed
-
- Alcami, A. 2003. Viral mimicry of cytokines, chemokines and their receptors. Nat. Rev. Immunol. 3:36-50. - PubMed
-
- Alcami, A., A. Khanna, N. L. Paul, and G. L. Smith. 1999. Vaccinia virus strains Lister, USSR and Evans express soluble and cell-surface tumour necrosis factor receptors. J. Gen. Virol. 80:949-959. - PubMed
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

