Modulation of the unfolded protein response by the severe acute respiratory syndrome coronavirus spike protein
- PMID: 16940539
- PMCID: PMC1563899
- DOI: 10.1128/JVI.00659-06
Modulation of the unfolded protein response by the severe acute respiratory syndrome coronavirus spike protein
Abstract
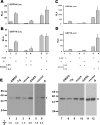
Perturbation of the function of endoplasmic reticulum (ER) causes stress leading to the activation of cell signaling pathways known as the unfolded protein response (UPR). Severe acute respiratory syndrome (SARS) coronavirus (SARS-CoV) uses ER as a site for synthesis and processing of viral proteins. In this report, we demonstrate that infection with SARS-CoV induces the UPR in cultured cells. A comparison with M, E, and NSP6 proteins indicates that SARS-CoV spike (S) protein sufficiently induces transcriptional activation of several UPR effectors, including glucose-regulated protein 78 (GRP78), GRP94, and C/EBP homologous protein. A substantial amount of S protein accumulates in the ER. The expression of S protein exerts different effects on the three major signaling pathways of the UPR. Particularly, it induces GRP78/94 through PKR-like ER kinase but has no influence on activating transcription factor 6 or X box-binding protein 1. Taken together, our findings suggest that SARS-CoV S protein specifically modulates the UPR to facilitate viral replication.
Figures
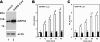



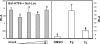
References
-
- Ball, J. M., T. Peng, C. Q. Y. Zeng, A. P. Morris, and M. K. Estes. 1996. Age-dependent diarrhea induced by a rotaviral nonstructural glycoprotein. Science 272:101-104. - PubMed
-
- Benali-Furet, N. L., M. Chami, L. Houel, F. De Giorgi, F. Vernejoul, D. Lagorce, L. Buscail, R. Bartenschlager, F. Ichas, R. Rizzuto, and P. Paterlini-Bréchot. 2005. Hepatitis C virus core triggers apoptosis in liver cells by inducing ER stress and ER calcium depletion. Oncogene 24:4921-4933. - PubMed
-
- Boyce, M., K. F. Bryant, C. Jousse, K. Long, H. P. Harding, D. Scheuner, R. J. Kaufman, D. Ma, D. M. Coen, D. Ron, and J. Yuan. 2005. A selective inhibitor of eIF2α dephosphorylation protects cells from ER stress. Science 307:935-939. - PubMed
-
- Boyce, M., and J. Yuan. 2006. Cellular response to endoplasmic reticulum stress: a matter of life or death. Cell Death Differ. 13:363-373. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

