Herpes simplex virus type 1 latently infected neurons differentially express latency-associated and ICP0 transcripts
- PMID: 16940542
- PMCID: PMC1563928
- DOI: 10.1128/JVI.02615-05
Herpes simplex virus type 1 latently infected neurons differentially express latency-associated and ICP0 transcripts
Abstract
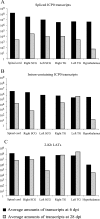
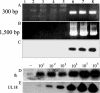
During the latent phase of herpes simplex virus type 1 (HSV-1) infection, the latency-associated transcripts (LATs) are the most abundant viral transcripts present in neurons, but some immediate-early viral transcripts, such as those encoding ICP0, have also been reported to be transcribed in latently infected mouse trigeminal ganglia (TG). A murine oro-ocular model of herpetic infection was used to study ICP0 gene expression in the major anatomical sites of HSV-1 latency, including the TG, superior cervical ganglion, spinal cord, and hypothalamus. An HSV-1 recombinant strain, SC16 110LacZ, revealed ICP0 promoter activity in several neurons in latently infected ganglia, and following infection with wild-type HSV-1 strain SC16, in situ hybridization analyses identified ICP0 transcripts in the nuclei of neurons at times consistent with the establishment of latency. Reverse transcription (RT)-PCR assays performed on RNA extracted from latently infected tissues indicated that ICP0 transcripts were detected in all anatomical sites of viral latency. Furthermore, quantitative real-time RT-PCR showed that neurons differentially expressed the LATs and ICP0 transcripts, with splicing of ICP0 transcripts being dependent on the anatomical location of latency. Finally, TG neurons were characterized by high-level expression of LATs and detection of abundant unspliced ICP0 transcripts, a pattern markedly different from those of other anatomical sites of HSV-1 latency. These results suggest that LATs might be involved in the maintenance of HSV-1 latency through the posttranscriptional regulation of ICP0 in order to inhibit expression of this potent activator of gene expression during latency.
Figures



References
-
- Arthur, J., S. Efstathiou, and A. Simmons. 1993. Intranuclear foci containing low abundance herpes simplex virus latency-associated transcripts visualized by non-isotopic in situ hybridization. J. Gen. Virol. 74:1363-1370. - PubMed
-
- Block, T. M., and J. M. Hill. 1997. The latency associated transcripts (LAT) of herpes simplex virus: still no end in sight. J. Neurovirol. 3:313-321. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

