DNA-mediated anisotropic mechanical reinforcement of a virus
- PMID: 16945903
- PMCID: PMC1564217
- DOI: 10.1073/pnas.0601881103
DNA-mediated anisotropic mechanical reinforcement of a virus
Abstract
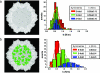
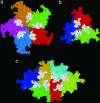
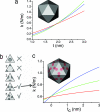
In this work, we provide evidence of a mechanism to reinforce the strength of an icosahedral virus by using its genomic DNA as a structural element. The mechanical properties of individual empty capsids and DNA-containing virions of the minute virus of mice are investigated by using atomic force microscopy. The stiffness of the empty capsid is found to be isotropic. Remarkably, the presence of the DNA inside the virion leads to an anisotropic reinforcement of the virus stiffness by approximately 3%, 40%, and 140% along the fivefold, threefold, and twofold symmetry axes, respectively. A finite element model of the virus indicates that this anisotropic mechanical reinforcement is due to DNA stretches bound to 60 concavities of the capsid. These results, together with evidence of biologically relevant conformational rearrangements of the capsid around pores located at the fivefold symmetry axes, suggest that the bound DNA may reinforce the overall stiffness of the viral particle without canceling the conformational changes needed for its infectivity.
Conflict of interest statement
Conflict of interest statement: No conflicts declared.
Figures

Similar articles
-
Structure of a packaging-defective mutant of minute virus of mice indicates that the genome is packaged via a pore at a 5-fold axis.J Virol. 2011 May;85(10):4822-7. doi: 10.1128/JVI.02598-10. Epub 2011 Mar 2. J Virol. 2011. PMID: 21367911 Free PMC article.
-
Imaging and Quantitation of a Succession of Transient Intermediates Reveal the Reversible Self-Assembly Pathway of a Simple Icosahedral Virus Capsid.J Am Chem Soc. 2016 Nov 30;138(47):15385-15396. doi: 10.1021/jacs.6b07663. Epub 2016 Nov 18. J Am Chem Soc. 2016. PMID: 27933931
-
Manipulation of the mechanical properties of a virus by protein engineering.Proc Natl Acad Sci U S A. 2008 Mar 18;105(11):4150-5. doi: 10.1073/pnas.0708017105. Epub 2008 Mar 11. Proc Natl Acad Sci U S A. 2008. PMID: 18334651 Free PMC article.
-
Structure, assembly, and DNA packaging of the bacteriophage T4 head.Adv Virus Res. 2012;82:119-53. doi: 10.1016/B978-0-12-394621-8.00018-2. Adv Virus Res. 2012. PMID: 22420853 Free PMC article. Review.
-
Condensed genome structure.Adv Exp Med Biol. 2012;726:469-87. doi: 10.1007/978-1-4614-0980-9_21. Adv Exp Med Biol. 2012. PMID: 22297527 Free PMC article. Review.
Cited by
-
Elucidating the mechanism behind irreversible deformation of viral capsids.Biophys J. 2009 Oct 7;97(7):2061-9. doi: 10.1016/j.bpj.2009.07.039. Biophys J. 2009. PMID: 19804738 Free PMC article.
-
Imaging modes of atomic force microscopy for application in molecular and cell biology.Nat Nanotechnol. 2017 Apr 6;12(4):295-307. doi: 10.1038/nnano.2017.45. Nat Nanotechnol. 2017. PMID: 28383040 Review.
-
Structural basis for biologically relevant mechanical stiffening of a virus capsid by cavity-creating or spacefilling mutations.Sci Rep. 2017 Jun 22;7(1):4101. doi: 10.1038/s41598-017-04345-w. Sci Rep. 2017. PMID: 28642465 Free PMC article.
-
Membrane-containing virus particles exhibit the mechanics of a composite material for genome protection.Nanoscale. 2018 Apr 26;10(16):7769-7779. doi: 10.1039/c8nr00196k. Nanoscale. 2018. PMID: 29658555 Free PMC article.
-
From viral assembly to host interaction: AFM's contributions to virology.J Virol. 2025 Jan 31;99(1):e0087324. doi: 10.1128/jvi.00873-24. Epub 2024 Dec 10. J Virol. 2025. PMID: 39655953 Free PMC article. Review.
References
-
- Hamm CE, Merkel R, Springer O, Jurkojc P, Maier C, Prechtel K, Smetacek V. Nature. 2003;421:841–843. - PubMed
-
- Goodman RP, Schaap IAT, Tardin CF, Erben CM, Berry RM, Schmidt CF, Turberfield AJ. Science. 2005;310:1661–1665. - PubMed
-
- Rossmann MG, Johnson JE. Annu Rev Biochem. 1989;58:533–573. - PubMed
-
- Johnson JE, Speir JA. In: Encyclopedia of Virology. Granoff A, Webster R, editors. London: Academic; 1999. pp. 1946–1956.
-
- Chiu W, Garcea RL, Burnett RM. Structural Biology of Viruses. Oxford: Oxford Univ Press; 1997.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

