NikR-operator complex structure and the mechanism of repressor activation by metal ions
- PMID: 16945905
- PMCID: PMC1564233
- DOI: 10.1073/pnas.0606247103
NikR-operator complex structure and the mechanism of repressor activation by metal ions
Abstract
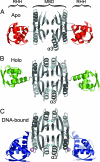
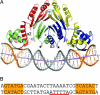
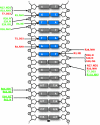
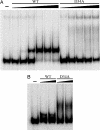
Metal ion homeostasis is critical to the survival of all cells. Regulation of nickel concentrations in Escherichia coli is mediated by the NikR repressor via nickel-induced transcriptional repression of the nickel ABC-type transporter, NikABCDE. Here, we report two crystal structures of nickel-activated E. coli NikR, the isolated repressor at 2.1 A resolution and in a complex with its operator DNA sequence from the nik promoter at 3.1 A resolution. Along with the previously published structure of apo-NikR, these structures allow us to evaluate functional proposals for how metal ions activate NikR, delineate the drastic conformational changes required for operator recognition, and describe the formation of a second metal-binding site in the presence of DNA. They also provide a rare set of structural views of a ligand-responsive transcription factor in the unbound, ligand-induced, and DNA-bound states, establishing a model system for the study of ligand-mediated effects on transcription factor function.
Conflict of interest statement
Conflict of interest statement: No conflicts declared.
Figures

References
-
- Mulrooney SB, Hausinger RP. FEMS Microbiol Rev. 2003;27:239–261. - PubMed
-
- Navarro C, Wu LF, Mandrand-Berthelot MA. Mol Microbiol. 1993;9:1181–1191. - PubMed
-
- Chivers PT, Sauer RT. J Biol Chem. 2000;275:19735–19741. - PubMed
-
- Contreras M, Thiberge JM, Mandrand-Berthelot MA, Labigne A. Mol Microbiol. 2003;49:947–963. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

