Hmgb3 regulates the balance between hematopoietic stem cell self-renewal and differentiation
- PMID: 16945912
- PMCID: PMC1564268
- DOI: 10.1073/pnas.0604006103
Hmgb3 regulates the balance between hematopoietic stem cell self-renewal and differentiation
Abstract
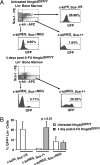
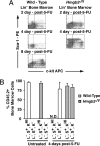
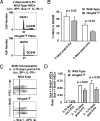
Hmgb3 is an X-linked member of a family of sequence-independent chromatin-binding proteins that is preferentially expressed in hematopoietic stem cells (HSC). Hmgb3-deficient mice (Hmgb3(-/Y)) contain normal numbers of HSCs, capable of self-renewal and hematopoietic repopulation, but fewer common lymphoid (CLP) and common myeloid progenitors (CMP). In this study, we tested the hypothesis that Hmgb3(-/Y) HSCs are biased toward self-renewal at the expense of progenitor production. Wild-type and Hmgb3(-/Y) CLPs and CMPs proliferate and differentiate equally in vitro, indicating that CLP and CMP function normally in Hmgb3(-/Y) mice. Hmgb3(-/Y) HSCs exhibit constitutive activation of the canonical Wnt signaling pathway, which regulates stem cell self-renewal. Increased Wnt signaling in Hmgb3(-/Y) HSCs corresponds to increased expression of Dvl1, a positive regulator of the canonical Wnt pathway. To induce hematopoietic stress and a subsequent response from HSCs, we treated Hmgb3(-/Y) mice with 5-fluorouracil. Hmgb3(-/Y) mice exhibit a faster recovery of functional HSCs after administration of 5-fluorouracil compared with wild-type mice, which may be due to the increased Wnt signaling. Furthermore, the recovery of HSC number in Hmgb3(-/Y) mice occurs more rapidly than CLP and CMP recovery. From these data, we propose a model in which Hmgb3 is required for the proper balance between HSC self-renewal and differentiation.
Conflict of interest statement
Conflict of interest statement: No conflicts declared.
Figures



Similar articles
-
Hmgb3 deficiency deregulates proliferation and differentiation of common lymphoid and myeloid progenitors.Blood. 2005 Jan 15;105(2):627-34. doi: 10.1182/blood-2004-07-2551. Epub 2004 Sep 9. Blood. 2005. PMID: 15358624
-
Hmgb3: an HMG-box family member expressed in primitive hematopoietic cells that inhibits myeloid and B-cell differentiation.Blood. 2003 Aug 15;102(4):1298-306. doi: 10.1182/blood-2002-11-3541. Epub 2003 Apr 24. Blood. 2003. PMID: 12714519
-
Lrp5 and Lrp6 are required for maintaining self-renewal and differentiation of hematopoietic stem cells.FASEB J. 2019 Apr;33(4):5615-5625. doi: 10.1096/fj.201802072R. Epub 2019 Jan 22. FASEB J. 2019. PMID: 30668923 Free PMC article.
-
Stem cell c-KIT and HOXB4 genes: critical roles and mechanisms in self-renewal, proliferation, and differentiation.Stem Cells Dev. 2006 Dec;15(6):755-78. doi: 10.1089/scd.2006.15.755. Stem Cells Dev. 2006. PMID: 17253940 Review.
-
Kit and Scl regulation of hematopoietic stem cells.Curr Opin Hematol. 2014 Jul;21(4):256-64. doi: 10.1097/MOH.0000000000000052. Curr Opin Hematol. 2014. PMID: 24857885 Review.
Cited by
-
The role of microRNAs in skeletal muscle health and disease.Front Biosci (Landmark Ed). 2015 Jan 1;20(1):37-77. doi: 10.2741/4298. Front Biosci (Landmark Ed). 2015. PMID: 25553440 Free PMC article. Review.
-
Construction and validation of a prognostic model based on 11 lymph node metastasis-related genes for overall survival in endometrial cancer.Cancer Med. 2022 Dec;11(23):4641-4655. doi: 10.1002/cam4.4844. Epub 2022 Jul 2. Cancer Med. 2022. PMID: 35778922 Free PMC article.
-
miRNA-mRNA integrative analysis in primary myelofibrosis CD34+ cells: role of miR-155/JARID2 axis in abnormal megakaryopoiesis.Blood. 2014 Sep 25;124(13):e21-32. doi: 10.1182/blood-2013-12-544197. Epub 2014 Aug 5. Blood. 2014. PMID: 25097177 Free PMC article.
-
Identification of disulfidptosis-related prognostic biomarkers associated with CD4 + and CD8 + T cells infiltration for sarcoma by integrating bioinformatic analysis and experimental validation.Discov Oncol. 2025 Jul 8;16(1):1284. doi: 10.1007/s12672-025-03127-5. Discov Oncol. 2025. PMID: 40627276 Free PMC article.
-
Prognostic value of HMGB3 expression in patients with non-small cell lung cancer.Tumour Biol. 2013 Oct;34(5):2599-603. doi: 10.1007/s13277-013-0807-y. Epub 2013 Apr 23. Tumour Biol. 2013. PMID: 23609034
References
-
- Kondo M, Wagers AJ, Manz MG, Prohaska SS, Scherer DC, Beilhack GF, Shizuru JA, Weissman IL. Annu Rev Immunol. 2003;21:759–806. - PubMed
-
- Bustin M, Lehn DA, Landsman D. Biochim Biophys Acta. 1990;1049:231–243. - PubMed
-
- Vaccari T, Beltrame M, Ferrari S, Bianchi M. Genomics. 1998;49:247–252. - PubMed
-
- Ronfani L, Ferraguti M, Croci L, Ovitt CE, Scholer HR, Consalez GG, Bianchi ME. Development (Cambridge, UK) 2001;128:1265–1273. - PubMed
-
- Bianchi ME, Beltrame M, Paonessa G. Science. 1989;243:1056–1059. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

