Inhibition of a spliceosome turnover pathway suppresses splicing defects
- PMID: 16945917
- PMCID: PMC1564266
- DOI: 10.1073/pnas.0603188103
Inhibition of a spliceosome turnover pathway suppresses splicing defects
Abstract
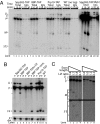
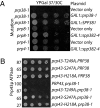
Defects in assembly are suggested to signal the dissociation of faulty splicing complexes. A yeast genetic screen was performed to identify components of the putative discard pathway. Weak mutant alleles of SPP382 (also called NTR1) were found to suppress defects in two proteins required for spliceosome activation, Prp38p and Prp8p. Spp382p is shown necessary for cellular splicing, with premRNA and, for some alleles, excised intron, accumulating after inactivation. Like spp382-1, a mutant allele of AAR2 was identified in this suppressor screen. Like Spp382p, Aar2p has a reported role in spliceosome recycling and is found with Spp382p in a complex recovered with a mutant version of the spliceosomal core protein Prp8p. Possible insight into to the spp382 suppressor phenotype is provided by the observation that defective splicing complexes lacking the 5' exon cleavage intermediate are recovered by a tandem affinity purification-tagged Spp382 derivative. Stringent proteomic and two-hybrid analyses show that Spp382p also interacts with Cwc23p, a DNA J-like protein present in the spliceosome and copurified with the Prp43p DExD/H-box ATPase. Spp382p binds Prp43p and Prp43p requires Spp382p for intron release from the spliceosome. Consistent with a related function in the removal of defective complexes, three prp43 mutants are also shown to suppress splicing defects, with efficiencies inversely proportionate to the measured ATPase activities. These and related genetic data support the existence of a Spp382p-dependent turnover pathway acting on defective spliceosomes.
Conflict of interest statement
Conflict of interest statement: No conflicts declared.
Figures

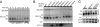
Similar articles
-
Spp382p interacts with multiple yeast splicing factors, including possible regulators of Prp43 DExD/H-Box protein function.Genetics. 2009 Sep;183(1):195-206. doi: 10.1534/genetics.109.106955. Epub 2009 Jul 6. Genetics. 2009. PMID: 19581443 Free PMC article.
-
Yeast ntr1/spp382 mediates prp43 function in postspliceosomes.Mol Cell Biol. 2006 Aug;26(16):6016-23. doi: 10.1128/MCB.02347-05. Mol Cell Biol. 2006. PMID: 16880513 Free PMC article.
-
The NineTeen Complex (NTC) and NTC-associated proteins as targets for spliceosomal ATPase action during pre-mRNA splicing.RNA Biol. 2015;12(2):109-14. doi: 10.1080/15476286.2015.1008926. RNA Biol. 2015. PMID: 25654271 Free PMC article.
-
Helicases involved in splicing from malaria parasite Plasmodium falciparum.Parasitol Int. 2011 Dec;60(4):335-40. doi: 10.1016/j.parint.2011.09.007. Epub 2011 Oct 1. Parasitol Int. 2011. PMID: 21996352 Review.
-
Functional roles of DExD/H-box RNA helicases in Pre-mRNA splicing.J Biomed Sci. 2015 Jul 16;22(1):54. doi: 10.1186/s12929-015-0161-z. J Biomed Sci. 2015. PMID: 26173448 Free PMC article. Review.
Cited by
-
Dissection of the factor requirements for spliceosome disassembly and the elucidation of its dissociation products using a purified splicing system.Genes Dev. 2013 Feb 15;27(4):413-28. doi: 10.1101/gad.207779.112. Genes Dev. 2013. PMID: 23431055 Free PMC article.
-
Structural basis for dual roles of Aar2p in U5 snRNP assembly.Genes Dev. 2013 Mar 1;27(5):525-40. doi: 10.1101/gad.213207.113. Epub 2013 Feb 26. Genes Dev. 2013. PMID: 23442228 Free PMC article.
-
Functional roles of protein splicing factors.Biosci Rep. 2012 Aug;32(4):345-59. doi: 10.1042/BSR20120007. Biosci Rep. 2012. PMID: 22762203 Free PMC article. Review.
-
Spliceosome discards intermediates via the DEAH box ATPase Prp43p.Proc Natl Acad Sci U S A. 2010 Jun 1;107(22):10020-5. doi: 10.1073/pnas.0906022107. Epub 2010 May 12. Proc Natl Acad Sci U S A. 2010. PMID: 20463285 Free PMC article.
-
Stalling of spliceosome assembly at distinct stages by small-molecule inhibitors of protein acetylation and deacetylation.RNA. 2009 Jan;15(1):153-75. doi: 10.1261/rna.1332609. Epub 2008 Nov 24. RNA. 2009. PMID: 19029308 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

