Structure of the stand-alone RAM-domain protein from Thermus thermophilus HB8
- PMID: 16946463
- PMCID: PMC2242884
- DOI: 10.1107/S1744309106031150
Structure of the stand-alone RAM-domain protein from Thermus thermophilus HB8
Abstract
The stand-alone RAM (regulation of amino-acid metabolism) domain protein SraA from Thermus thermophilus HB8 (TTHA0845) was crystallized in the presence of zinc ions. The X-ray crystal structure was determined using a multiple-wavelength anomalous dispersion technique and was refined at 2.4 A resolution to a final R factor of 25.0%. The monomeric structure is a betaalphabetabetaalphabeta fold and it dimerizes mainly through interactions between the antiparallel beta-sheets. Furthermore, five SraA dimers form a ring with external and internal diameters of 70 and 20 A, respectively. This decameric structure is unique compared with the octameric and dodecameric structures found for other stand-alone RAM-domain proteins and the C-terminal RAM domains of Lrp/AsnC-family proteins.
Figures

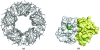
Similar articles
-
Novel stand-alone RAM domain protein-mediated catalytic control of anthranilate phosphoribosyltransferase in tryptophan biosynthesis in Thermus thermophilus.Extremophiles. 2017 Jan;21(1):73-83. doi: 10.1007/s00792-016-0884-0. Epub 2016 Oct 19. Extremophiles. 2017. PMID: 27757697
-
Crystal structure of uncharacterized protein TTHA1756 from Thermus thermophilus HB8: Structural variety in UPF0150 family proteins.Proteins. 2008 Jun;71(4):2097-101. doi: 10.1002/prot.22018. Proteins. 2008. PMID: 18338381 No abstract available.
-
Crystal structure of a predicted phosphoribosyltransferase (TT1426) from Thermus thermophilus HB8 at 2.01 A resolution.Protein Sci. 2005 Mar;14(3):823-7. doi: 10.1110/ps.041229405. Epub 2005 Feb 2. Protein Sci. 2005. PMID: 15689504 Free PMC article.
-
Bioenergetics at extreme temperature: Thermus thermophilus ba(3)- and caa(3)-type cytochrome c oxidases.Biochim Biophys Acta. 2012 Apr;1817(4):638-49. doi: 10.1016/j.bbabio.2011.08.004. Biochim Biophys Acta. 2012. PMID: 22385645 Review.
-
Hyperthermophiles: taking the heat and loving it.Structure. 1995 Mar 15;3(3):251-4. doi: 10.1016/s0969-2126(01)00155-1. Structure. 1995. PMID: 7788291 Review. No abstract available.
Cited by
-
Novel stand-alone RAM domain protein-mediated catalytic control of anthranilate phosphoribosyltransferase in tryptophan biosynthesis in Thermus thermophilus.Extremophiles. 2017 Jan;21(1):73-83. doi: 10.1007/s00792-016-0884-0. Epub 2016 Oct 19. Extremophiles. 2017. PMID: 27757697
References
-
- Bell, S. D. & Jackson, S. P. (2001). Curr. Opin. Microbiol.4, 208–213. - PubMed
-
- Brinkman, A. B., Ettema, T. J., de Vos, W. M. & van der Oost, J. (2003). Mol. Microbiol.48, 287–294. - PubMed
-
- Brünger, A. T., Adams, P. D., Clore, G. M., DeLano, W. L., Gros, P., Grosse-Kunstleve, R. W., Jiang, J.-S., Kuszewski, J., Nilges, M., Pannu, N. S., Read, R. J., Rice, L. M., Simonson, T. & Warren, G. L. (1998). Acta Cryst. D54, 905–921. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Miscellaneous

