3'-end formation signals modulate the association of genes with the nuclear periphery as well as mRNP dot formation
- PMID: 16946703
- PMCID: PMC1570430
- DOI: 10.1038/sj.emboj.7601305
3'-end formation signals modulate the association of genes with the nuclear periphery as well as mRNP dot formation
Abstract
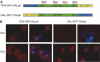
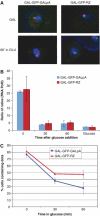
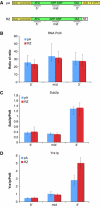
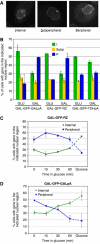
Multiple studies indicate that mRNA processing defects cause mRNAs to accumulate in discrete nuclear foci or dots, in mammalian cells as well as yeast. To investigate this phenomenon, we have studied a series of GAL reporter constructs integrated into the yeast genome adjacent to an array of TetR-GFP-bound TetO sites. mRNA within dots is predominantly post-transcriptional, and dots are adjacent to but distinct from their transcription site. These reporter genes also localize to the nuclear periphery upon gene induction, like their endogenous GAL counterparts. Surprisingly, this peripheral localization persists long after transcriptional shutoff, and there is a comparable persistence of the RNA in the dots. Moreover, dot disappearance and gene delocalization from the nuclear periphery occur with similar kinetics after transcriptional shutoff. Both kinetics depend in turn on reporter gene 3'-end formation signals. Our experiments indicate that gene association with the nuclear periphery does not require ongoing transcription and suggest that the mRNPs within dots may make a major contribution to the gene-nuclear periphery tether.
Figures



References
-
- Agard DA, Hiraoka Y, Shaw P, Sedat JW (1989) Fluorescence microscopy in three dimensions. Methods Cell Biol 30: 353–377 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

