Mechanism of RecA-mediated homologous recombination revisited by single molecule nanomanipulation
- PMID: 16946710
- PMCID: PMC1570433
- DOI: 10.1038/sj.emboj.7601260
Mechanism of RecA-mediated homologous recombination revisited by single molecule nanomanipulation
Abstract
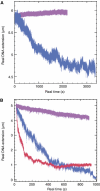
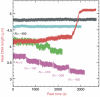
The mechanisms of RecA-mediated three-strand homologous recombination are investigated at the single-molecule level, using magnetic tweezers. Probing the mechanical response of DNA molecules and nucleoprotein filaments in tension and in torsion allows a monitoring of the progression of the exchange in real time, both from the point of view of the RecA-bound single-stranded DNA and from that of the naked double-stranded DNA (dsDNA). We show that strand exchange is able to generate torsion even along a molecule with freely rotating ends. RecA readily depolymerizes during the reaction, a process presenting numerous advantages for the cell's 'protein economy' and for the management of topological constraints. Invasion of an untwisted dsDNA by a nucleoprotein filament leads to an exchanged duplex that remains topologically linked to the exchanged single strand, suggesting multiple initiations of strand exchange on the same molecule. Overall, our results seem to support several important assumptions of the monomer redistribution model.
Figures




Similar articles
-
Elucidating a key intermediate in homologous DNA strand exchange: structural characterization of the RecA-triple-stranded DNA complex using fluorescence resonance energy transfer.J Mol Biol. 2002 Jul 12;320(3):529-58. doi: 10.1016/s0022-2836(02)00462-x. J Mol Biol. 2002. PMID: 12096908
-
Homologous recombination in real time: DNA strand exchange by RecA.Mol Cell. 2008 May 23;30(4):530-8. doi: 10.1016/j.molcel.2008.03.010. Mol Cell. 2008. PMID: 18498754
-
ATP hydrolysis Promotes Duplex DNA Release by the RecA Presynaptic Complex.J Biol Chem. 2016 Oct 14;291(42):22218-22230. doi: 10.1074/jbc.M116.740563. Epub 2016 Sep 1. J Biol Chem. 2016. PMID: 27587394 Free PMC article.
-
Structure of RecA-DNA complex and mechanism of DNA strand exchange reaction in homologous recombination.Adv Biophys. 1994;30:1-35. doi: 10.1016/0065-227x(94)90009-4. Adv Biophys. 1994. PMID: 7709802 Review.
-
RecA and DNA recombination: a review of molecular mechanisms.Biochem Soc Trans. 2019 Oct 31;47(5):1511-1531. doi: 10.1042/BST20190558. Biochem Soc Trans. 2019. PMID: 31654073 Review.
Cited by
-
Mismatch detection in homologous strand exchange amplified by hydrophobic effects.Biopolymers. 2021 Apr;112(4):e23426. doi: 10.1002/bip.23426. Epub 2021 Mar 29. Biopolymers. 2021. PMID: 33780001 Free PMC article.
-
Single-molecule insight into stalled replication fork rescue in Escherichia coli.Nucleic Acids Res. 2021 May 7;49(8):4220-4238. doi: 10.1093/nar/gkab142. Nucleic Acids Res. 2021. PMID: 33744948 Free PMC article. Review.
-
An Overview of the Molecular Mechanisms of Recombinational DNA Repair.Cold Spring Harb Perspect Biol. 2015 Nov 2;7(11):a016410. doi: 10.1101/cshperspect.a016410. Cold Spring Harb Perspect Biol. 2015. PMID: 26525148 Free PMC article. Review.
-
Multi-scale tracking reveals scale-dependent chromatin dynamics after DNA damage.Mol Biol Cell. 2017 Aug 9;28(23):3323-32. doi: 10.1091/mbc.E17-05-0317. Online ahead of print. Mol Biol Cell. 2017. PMID: 28794266 Free PMC article.
-
Single molecule studies of homologous recombination.Mol Biosyst. 2008 Nov;4(11):1094-104. doi: 10.1039/b811681b. Epub 2008 Sep 29. Mol Biosyst. 2008. PMID: 18931785 Free PMC article. Review.
References
-
- Adzuma K (1992) Stable synapsis of homologous DNA molecules mediated by the Escherichia coli RecA protein involves local exchange of DNA strands. Gene Dev 6: 1679–1694 - PubMed
-
- Bennink ML, Scharer OD, Kanaar R, Sakata-Sogawa K, Schins JM, Kanger JS, de Grooth BG, Greve J (1999) Single-molecule manipulation of double-stranded DNA using optical tweezers: interaction studies of DNA with RecA and YOYO-1. Cytometry 36: 200–208 - PubMed
-
- Bianco PR, Tracy RB, Kowalczykowski SC (1998) DNA strand exchange proteins: a biochemical and physical comparison. Front Biosci 3: D570–D603 - PubMed
-
- Bugreeva I, Bugreev D, Nevinsky G (2005) Formation of nucleoprotein RecA filament on single-stranded DNA. FEBS J 272: 2734–2745 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

