The human urinary proteome contains more than 1500 proteins, including a large proportion of membrane proteins
- PMID: 16948836
- PMCID: PMC1794545
- DOI: 10.1186/gb-2006-7-9-R80
The human urinary proteome contains more than 1500 proteins, including a large proportion of membrane proteins
Abstract
Background: Urine is a desirable material for the diagnosis and classification of diseases because of the convenience of its collection in large amounts; however, all of the urinary proteome catalogs currently being generated have limitations in their depth and confidence of identification. Our laboratory has developed methods for the in-depth characterization of body fluids; these involve a linear ion trap-Fourier transform (LTQ-FT) and a linear ion trap-orbitrap (LTQ-Orbitrap) mass spectrometer. Here we applied these methods to the analysis of the human urinary proteome.
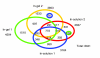
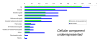
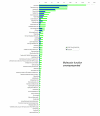
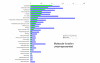
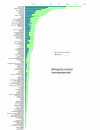
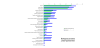
Results: We employed one-dimensional sodium dodecyl sulfate polyacrylamide gel electrophoresis and reverse phase high-performance liquid chromatography for protein separation and fractionation. Fractionated proteins were digested in-gel or in-solution, and digests were analyzed with the LTQ-FT and LTQ-Orbitrap at parts per million accuracy and with two consecutive stages of mass spectrometric fragmentation. We identified 1543 proteins in urine obtained from ten healthy donors, while essentially eliminating false-positive identifications. Surprisingly, nearly half of the annotated proteins were membrane proteins according to Gene Ontology (GO) analysis. Furthermore, extracellular, lysosomal, and plasma membrane proteins were enriched in the urine compared with all GO entries. Plasma membrane proteins are probably present in urine by secretion in exosomes.
Conclusion: Our analysis provides a high-confidence set of proteins present in human urinary proteome and provides a useful reference for comparing datasets obtained using different methodologies. The urinary proteome is unexpectedly complex and may prove useful in biomarker discovery in the future.
Figures






References
-
- Brenner B, (editor) The Kidney. Philadelphia, PA: WB Saunders; 2000.
-
- Brunzel NA. Fundamentals of Urine & Body Fluid Analysis. Philadelphia, PA: Saunders; 2004.
-
- Maunsbach AB. Absorption of I125-labeled homologous albumin by rat kidney proximal tubule cells. A study of microperfused single proximal tubules by electron microscopic autoradiography and histochemistry. 1966. J Am Soc Nephrol. 1997;8:323–351. discussion 327-331. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

