The complete sequences and gene organisation of the mitochondrial genomes of the heterodont bivalves Acanthocardia tuberculata and Hiatella arctica--and the first record for a putative Atpase subunit 8 gene in marine bivalves
- PMID: 16948842
- PMCID: PMC1570459
- DOI: 10.1186/1742-9994-3-13
The complete sequences and gene organisation of the mitochondrial genomes of the heterodont bivalves Acanthocardia tuberculata and Hiatella arctica--and the first record for a putative Atpase subunit 8 gene in marine bivalves
Abstract
Background: Mitochondrial (mt) gene arrangement is highly variable among molluscs and especially among bivalves. Of the 30 complete molluscan mt-genomes published to date, only one is of a heterodont bivalve, although this is the most diverse taxon in terms of species numbers. We determined the complete sequence of the mitochondrial genomes of Acanthocardia tuberculata and Hiatella arctica, (Mollusca, Bivalvia, Heterodonta) and describe their gene contents and genome organisations to assess the variability of these features among the Bivalvia and their value for phylogenetic inference.
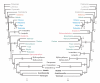
Results: The size of the mt-genome in Acanthocardia tuberculata is 16.104 basepairs (bp), and in Hiatella arctica 18.244 bp. The Acanthocardia mt-genome contains 12 of the typical protein coding genes, lacking the Atpase subunit 8 (atp8) gene, as all published marine bivalves. In contrast, a complete atp8 gene is present in Hiatella arctica. In addition, we found a putative truncated atp8 gene when re-annotating the mt-genome of Venerupis philippinarum. Both mt-genomes reported here encode all genes on the same strand and have an additional trnM. In Acanthocardia several large non-coding regions are present. One of these contains 3.5 nearly identical copies of a 167 bp motive. In Hiatella, the 3' end of the NADH dehydrogenase subunit (nad)6 gene is duplicated together with the adjacent non-coding region. The gene arrangement of Hiatella is markedly different from all other known molluscan mt-genomes, that of Acanthocardia shows few identities with the Venerupis philippinarum. Phylogenetic analyses on amino acid and nucleotide levels robustly support the Heterodonta and the sister group relationship of Acanthocardia and Venerupis. Monophyletic Bivalvia are resolved only by a Bayesian inference of the nucleotide data set. In all other analyses the two unionid species, being to only ones with genes located on both strands, do not group with the remaining bivalves.
Conclusion: The two mt-genomes reported here add to and underline the high variability of gene order and presence of duplications in bivalve and molluscan taxa. Some genomic traits like the loss of the atp8 gene or the encoding of all genes on the same strand are homoplastic among the Bivalvia. These characters, gene order, and the nucleotide sequence data show considerable potential of resolving phylogenetic patterns at lower taxonomic levels.
Figures




References
-
- Wolstenholme DR. Animal mitochondrial DNA: Structure and evolution. Int Rev Cytol. 1992;141:173–216. - PubMed
-
- Boore JL, Staton JL. The mitochondrial genome of the sipunculid Phascolopsis gouldii supports its association with Annelida rather than Mollusca. Mol Biol Evol. 2002;19:127–137. - PubMed
-
- Stewart DT, Saavedra C, Stanwood RR, Ball AO, Zouros E. Male and female mitochondrial DNA lineages in the Blue Mussel Mytilus. Mol Biol Evol. 1995;12:735–747. - PubMed
LinkOut - more resources
Full Text Sources

