PhyloPat: phylogenetic pattern analysis of eukaryotic genes
- PMID: 16948844
- PMCID: PMC1570148
- DOI: 10.1186/1471-2105-7-398
PhyloPat: phylogenetic pattern analysis of eukaryotic genes
Abstract
Background: Phylogenetic patterns show the presence or absence of certain genes or proteins in a set of species. They can also be used to determine sets of genes or proteins that occur only in certain evolutionary branches. Phylogenetic patterns analysis has routinely been applied to protein databases such as COG and OrthoMCL, but not upon gene databases. Here we present a tool named PhyloPat which allows the complete Ensembl gene database to be queried using phylogenetic patterns.
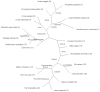
Description: PhyloPat is an easy-to-use webserver, which can be used to query the orthologies of all complete genomes within the EnsMart database using phylogenetic patterns. This enables the determination of sets of genes that occur only in certain evolutionary branches or even single species. We found in total 446,825 genes and 3,164,088 orthologous relationships within the EnsMart v40 database. We used a single linkage clustering algorithm to create 147,922 phylogenetic lineages, using every one of the orthologies provided by Ensembl. PhyloPat provides the possibility of querying with either binary phylogenetic patterns (created by checkboxes) or regular expressions. Specific branches of a phylogenetic tree of the 21 included species can be selected to create a branch-specific phylogenetic pattern. Users can also input a list of Ensembl or EMBL IDs to check which phylogenetic lineage any gene belongs to. The output can be saved in HTML, Excel or plain text format for further analysis. A link to the FatiGO web interface has been incorporated in the HTML output, creating easy access to functional information. Finally, lists of omnipresent, polypresent and oligopresent genes have been included.
Conclusion: PhyloPat is the first tool to combine complete genome information with phylogenetic pattern querying. Since we used the orthologies generated by the accurate pipeline of Ensembl, the obtained phylogenetic lineages are reliable. The completeness and reliability of these phylogenetic lineages will further increase with the addition of newly found orthologous relationships within each new Ensembl release.
Figures



References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

