Cancer diagnosis marker extraction for soft tissue sarcomas based on gene expression profiling data by using projective adaptive resonance theory (PART) filtering method
- PMID: 16948864
- PMCID: PMC1569882
- DOI: 10.1186/1471-2105-7-399
Cancer diagnosis marker extraction for soft tissue sarcomas based on gene expression profiling data by using projective adaptive resonance theory (PART) filtering method
Abstract
Background: Recent advances in genome technologies have provided an excellent opportunity to determine the complete biological characteristics of neoplastic tissues, resulting in improved diagnosis and selection of treatment. To accomplish this objective, it is important to establish a sophisticated algorithm that can deal with large quantities of data such as gene expression profiles obtained by DNA microarray analysis.
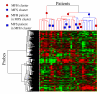
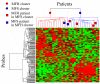
Results: Previously, we developed the projective adaptive resonance theory (PART) filtering method as a gene filtering method. This is one of the clustering methods that can select specific genes for each subtype. In this study, we applied the PART filtering method to analyze microarray data that were obtained from soft tissue sarcoma (STS) patients for the extraction of subtype-specific genes. The performance of the filtering method was evaluated by comparison with other widely used methods, such as signal-to-noise, significance analysis of microarrays, and nearest shrunken centroids. In addition, various combinations of filtering and modeling methods were used to extract essential subtype-specific genes. The combination of the PART filtering method and boosting--the PART-BFCS method--showed the highest accuracy. Seven genes among the 15 genes that are frequently selected by this method--MIF, CYFIP2, HSPCB, TIMP3, LDHA, ABR, and RGS3--are known prognostic marker genes for other tumors. These genes are candidate marker genes for the diagnosis of STS. Correlation analysis was performed to extract marker genes that were not selected by PART-BFCS. Sixteen genes among those extracted are also known prognostic marker genes for other tumors, and they could be candidate marker genes for the diagnosis of STS.
Conclusion: The procedure that consisted of two steps, such as the PART-BFCS and the correlation analysis, was proposed. The results suggest that novel diagnostic and therapeutic targets for STS can be extracted by a procedure that includes the PART filtering method.
Figures

Similar articles
-
Macrophage migration inhibitory factor and stearoyl-CoA desaturase 1: potential prognostic markers for soft tissue sarcomas based on bioinformatics analyses.PLoS One. 2013 Oct 22;8(10):e78250. doi: 10.1371/journal.pone.0078250. eCollection 2013. PLoS One. 2013. PMID: 24167613 Free PMC article. Clinical Trial.
-
Analysis of gene expression profiles of soft tissue sarcoma using a combination of knowledge-based filtering with integration of multiple statistics.PLoS One. 2014 Sep 4;9(9):e106801. doi: 10.1371/journal.pone.0106801. eCollection 2014. PLoS One. 2014. PMID: 25188299 Free PMC article.
-
Modified signal-to-noise: a new simple and practical gene filtering approach based on the concept of projective adaptive resonance theory (PART) filtering method.Bioinformatics. 2006 Jul 1;22(13):1662-4. doi: 10.1093/bioinformatics/btl156. Epub 2006 Apr 21. Bioinformatics. 2006. PMID: 16632489
-
Lessons from genetic profiling in soft tissue sarcomas.Acta Orthop Scand Suppl. 2004 Apr;75(311):35-50. doi: 10.1080/00016470410001708310. Acta Orthop Scand Suppl. 2004. PMID: 15188664 Review.
-
Molecular profiling in the diagnosis and treatment of high grade sarcomas.Ultrastruct Pathol. 2008 Mar-Apr;32(2):37-42. doi: 10.1080/01913120801897174. Ultrastruct Pathol. 2008. PMID: 18446666 Review.
Cited by
-
Cyfip2 controls the acoustic startle threshold through FMRP, actin polymerization, and GABAB receptor function.bioRxiv [Preprint]. 2024 Feb 5:2023.12.22.573054. doi: 10.1101/2023.12.22.573054. bioRxiv. 2024. PMID: 38187577 Free PMC article. Preprint.
-
Genetic variations in the regulator of G-protein signaling genes are associated with survival in late-stage non-small cell lung cancer.PLoS One. 2011;6(6):e21120. doi: 10.1371/journal.pone.0021120. Epub 2011 Jun 17. PLoS One. 2011. PMID: 21698121 Free PMC article.
-
Construction of possible integrated predictive index based on EGFR and ANXA3 polymorphisms for chemotherapy response in fluoropyrimidine-treated Japanese gastric cancer patients using a bioinformatic method.BMC Cancer. 2015 Oct 16;15:718. doi: 10.1186/s12885-015-1721-z. BMC Cancer. 2015. PMID: 26475168 Free PMC article.
-
Macrophage migration inhibitory factor and stearoyl-CoA desaturase 1: potential prognostic markers for soft tissue sarcomas based on bioinformatics analyses.PLoS One. 2013 Oct 22;8(10):e78250. doi: 10.1371/journal.pone.0078250. eCollection 2013. PLoS One. 2013. PMID: 24167613 Free PMC article. Clinical Trial.
-
Accurate molecular characterization of formalin-fixed, paraffin-embedded tissues by microRNA expression profiling.J Mol Diagn. 2008 Sep;10(5):415-23. doi: 10.2353/jmoldx.2008.080018. Epub 2008 Aug 7. J Mol Diagn. 2008. PMID: 18687792 Free PMC article.
References
-
- Golub TR, Slonim DK, Tamayo P, Huard C, Gaasenbeek M, Mesirov JP, Coller H, Loh ML, Downing JR, Caligiuri MA, Bloomfield CD, Lander ES. Molecular classification of cancer: class discovery and class prediction by gene expression monitoring. Science. 1999;286:531–537. doi: 10.1126/science.286.5439.531. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

