Co-evolutionary analysis of domains in interacting proteins reveals insights into domain-domain interactions mediating protein-protein interactions
- PMID: 16949097
- PMCID: PMC1618801
- DOI: 10.1016/j.jmb.2006.07.072
Co-evolutionary analysis of domains in interacting proteins reveals insights into domain-domain interactions mediating protein-protein interactions
Abstract
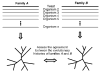
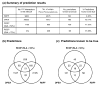
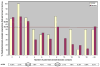
Recent advances in functional genomics have helped generate large-scale high-throughput protein interaction data. Such networks, though extremely valuable towards molecular level understanding of cells, do not provide any direct information about the regions (domains) in the proteins that mediate the interaction. Here, we performed co-evolutionary analysis of domains in interacting proteins in order to understand the degree of co-evolution of interacting and non-interacting domains. Using a combination of sequence and structural analysis, we analyzed protein-protein interactions in F1-ATPase, Sec23p/Sec24p, DNA-directed RNA polymerase and nuclear pore complexes, and found that interacting domain pair(s) for a given interaction exhibits higher level of co-evolution than the non-interacting domain pairs. Motivated by this finding, we developed a computational method to test the generality of the observed trend, and to predict large-scale domain-domain interactions. Given a protein-protein interaction, the proposed method predicts the domain pair(s) that is most likely to mediate the protein interaction. We applied this method on the yeast interactome to predict domain-domain interactions, and used known domain-domain interactions found in PDB crystal structures to validate our predictions. Our results show that the prediction accuracy of the proposed method is statistically significant. Comparison of our prediction results with those from two other methods reveals that only a fraction of predictions are shared by all the three methods, indicating that the proposed method can detect known interactions missed by other methods. We believe that the proposed method can be used with other methods to help identify previously unrecognized domain-domain interactions on a genome scale, and could potentially help reduce the search space for identifying interaction sites.
Figures




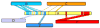

References
-
- Uetz P, Giot L, Cagney G, Mansfield TA, Judson RS, Knight JR, et al. A comprehensive analysis of protein–protein interactions in Saccharomyces cerevisiae. Nature. 2000;403:623–627. - PubMed
-
- Gavin AC, Bosche M, Krause R, Grandi P, Marzioch M, Bauer A, et al. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature. 2002;415:141–147. - PubMed
-
- Ho Y, Gruhler A, Heilbut A, Bader GD, Moore L, Adams SL, et al. Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature. 2002;415:180–183. - PubMed
-
- Giot L, Bader JS, Brouwer C, Chaudhuri A, Kuang B, Li Y, et al. A protein interaction map of Drosophila melanogaster. Science. 2003;302:1727–1736. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

