Diffraction-based density restraints for membrane and membrane-peptide molecular dynamics simulations
- PMID: 16950837
- PMCID: PMC1630481
- DOI: 10.1529/biophysj.106.084483
Diffraction-based density restraints for membrane and membrane-peptide molecular dynamics simulations
Abstract
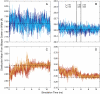
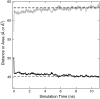
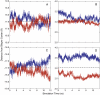
We have recently shown that current molecular dynamics (MD) atomic force fields are not yet able to produce lipid bilayer structures that agree with experimentally-determined structures within experimental errors. Because of the many advantages offered by experimentally validated simulations, we have developed a novel restraint method for membrane MD simulations that uses experimental diffraction data. The restraints, introduced into the MD force field, act upon specified groups of atoms to restrain their mean positions and widths to values determined experimentally. The method was first tested using a simple liquid argon system, and then applied to a neat dioleoylphosphatidylcholine (DOPC) bilayer at 66% relative humidity and to the same bilayer containing the peptide melittin. Application of experiment-based restraints to the transbilayer double-bond and water distributions of neat DOPC bilayers led to distributions that agreed with the experimental values. Based upon the experimental structure, the restraints improved the simulated structure in some regions while introducing larger differences in others, as might be expected from imperfect force fields. For the DOPC-melittin system, the experimental transbilayer distribution of melittin was used as a restraint. The addition of the peptide caused perturbations of the simulated bilayer structure, but which were larger than observed experimentally. The melittin distribution of the simulation could be fit accurately to a Gaussian with parameters close to the observed ones, indicating that the restraints can be used to produce an ensemble of membrane-bound peptide conformations that are consistent with experiments. Such ensembles pave the way for understanding peptide-bilayer interactions at the atomic level.
Figures
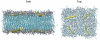








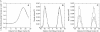
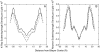
Similar articles
-
Conformational states of melittin at a bilayer interface.Biophys J. 2013 Mar 19;104(6):L12-4. doi: 10.1016/j.bpj.2013.02.006. Epub 2013 Mar 19. Biophys J. 2013. PMID: 23528098 Free PMC article.
-
Experimental validation of molecular dynamics simulations of lipid bilayers: a new approach.Biophys J. 2005 Feb;88(2):805-17. doi: 10.1529/biophysj.104.046821. Epub 2004 Nov 8. Biophys J. 2005. PMID: 15533925 Free PMC article.
-
Binding and reorientation of melittin in a POPC bilayer: computer simulations.Biochim Biophys Acta. 2012 Dec;1818(12):2975-81. doi: 10.1016/j.bbamem.2012.07.026. Epub 2012 Aug 2. Biochim Biophys Acta. 2012. PMID: 22877705
-
Simulation studies of the interaction of antimicrobial peptides and lipid bilayers.Biochim Biophys Acta. 1999 Dec 15;1462(1-2):185-200. doi: 10.1016/s0005-2736(99)00206-0. Biochim Biophys Acta. 1999. PMID: 10590308 Review.
-
Molecular dynamics simulation studies of lipid bilayer systems.Acta Biochim Pol. 2000;47(3):601-11. Acta Biochim Pol. 2000. PMID: 11310963 Review.
Cited by
-
Membrane association of the PTEN tumor suppressor: neutron scattering and MD simulations reveal the structure of protein-membrane complexes.Methods. 2015 May;77-78:136-46. doi: 10.1016/j.ymeth.2014.10.014. Epub 2014 Oct 27. Methods. 2015. PMID: 25461777 Free PMC article. Review.
-
The interaction of phospholipase A2 with a phospholipid bilayer: coarse-grained molecular dynamics simulations.Biophys J. 2008 Aug;95(4):1649-57. doi: 10.1529/biophysj.107.123190. Epub 2008 May 9. Biophys J. 2008. PMID: 18469074 Free PMC article.
-
Conformational states of melittin at a bilayer interface.Biophys J. 2013 Mar 19;104(6):L12-4. doi: 10.1016/j.bpj.2013.02.006. Epub 2013 Mar 19. Biophys J. 2013. PMID: 23528098 Free PMC article.
-
Membrane protein insertion: the biology-physics nexus.J Gen Physiol. 2007 May;129(5):363-9. doi: 10.1085/jgp.200709741. Epub 2007 Apr 16. J Gen Physiol. 2007. PMID: 17438116 Free PMC article. Review. No abstract available.
-
Zooming in on disordered systems: neutron reflection studies of proteins associated with fluid membranes.Biochim Biophys Acta. 2014 Sep;1838(9):2341-9. doi: 10.1016/j.bbamem.2014.03.007. Epub 2014 Mar 25. Biochim Biophys Acta. 2014. PMID: 24674984 Free PMC article. Review.
References
-
- McIntosh, T. J. 1990. X-ray diffraction analysis of membrane lipids. In Molecular Description of Biological Membrane by Computer Aided Conformational Analysis. R. Brasseur, editor. CRC Press. Boca Raton, FL. 241–266.
-
- White, S. H., and M. C. Wiener. 1995. Determination of the structure of fluid lipid bilayer membranes. In Permeability and Stability of Lipid Bilayers. E. A. Disalvo and S. A. Simon, editors. CRC Press. Boca Raton, FL. 1–19.
-
- Franks, N. P., and Y. K. Levine. 1981. Low-angle X-ray diffraction. In Membrane Spectroscopy. E. Grell, editor. Springer-Verlag. Berlin, Germany. 437–487.

