Advantage of using inosine at the 3' termini of 16S rRNA gene universal primers for the study of microbial diversity
- PMID: 16950904
- PMCID: PMC1636166
- DOI: 10.1128/AEM.00849-06
Advantage of using inosine at the 3' termini of 16S rRNA gene universal primers for the study of microbial diversity
Abstract
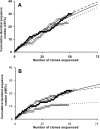
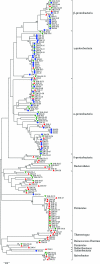
To overcome the shortcomings of universal 16S rRNA gene primers 8F and 907R when studying the diversity of complex microbial communities, the 3' termini of both primers were replaced with inosine. A comparison of the clone libraries derived using both primer sets showed seven bacterial phyla amplified by the altered primer set (8F-I/907R-I) whereas the original set amplified sequences belonging almost exclusively to Proteobacteria (95.8%). Sequences belonging to Firmicutes (42.6%) and Thermotogae (9.3%) were more abundant in a library obtained by using 8F-I/907R-I at a PCR annealing temperature of 54 degrees C, while Proteobacteria sequences were more frequent (62.7%) in a library obtained at 50 degrees C, somewhat resembling the result obtained using the original primer set. The increased diversity revealed by using primers 8F-I/907R-I confirms the usefulness of primers with inosine at the 3' termini in studying the microbial diversity of environmental samples.
Figures
References
-
- Baker, G. C., J. J. Smith, and D. A. Cowan. 2003. Review and re-analysis of domain-specific 16S primers. J. Microbiol. Methods 55:541-555. - PubMed
-
- Crick, F. H. C. 1966. Codon-anticodon pairing: the wobble hypothesis. J. Mol. Biol. 19:548-555. - PubMed
-
- Daly, K., R. J. Sharp, and A. J. McCarthy. 2000. Development of oligonucleotide probes and PCR primers for detecting phylogenetic subgroups of sulfate-reducing bacteria. Microbiology 146:1693-1705. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

