Interaction between coat morphogenetic proteins SafA and SpoVID
- PMID: 16950916
- PMCID: PMC1636312
- DOI: 10.1128/JB.00761-06
Interaction between coat morphogenetic proteins SafA and SpoVID
Abstract
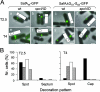
Morphogenetic proteins such as SpoVID and SafA govern assembly of the Bacillus subtilis endospore coat by guiding the various protein structural components to the surface of the developing spore. Previously, a screen for peptides able to interact with SpoVID led to the identification of a PYYH motif present in the C-terminal half of the SafA protein and to the subsequent demonstration that SpoVID and SafA directly interact. spoVID and safA spores show deficiencies in coat assembly and are lysozyme susceptible. Both proteins, orthologs of which are found in all Bacillus species, have LysM domains for peptidoglycan binding and localize to the cortex-coat interface. Here, we show that the interaction between SafA and SpoVID involves the PYYH motif (region B) but also a 13-amino-acid region (region A) just downstream of the N-terminal LysM domain of SafA. We show that deletion of region B does not block the interaction of SafA with SpoVID, nor does it bring about spore susceptibility to lysozyme. Nevertheless, it appears to reduce the interaction and affects the complex. In contrast, lesions in region A impaired the interaction of SafA with SpoVID in vitro and, while not affecting the accumulation of SafA in vivo, interfered with the localization of SafA around the developing spore, causing aberrant assembly of the coat and lysozyme sensitivity. A peptide corresponding to region A interacts with SpoVID, suggesting that residues within this region directly contact SpoVID. Since region A is highly conserved among SafA orthologs, this motif may be an important determinant of coat assembly in the group of Bacillus spore formers.
Figures




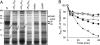
Similar articles
-
Morphogenetic proteins SpoVID and SafA form a complex during assembly of the Bacillus subtilis spore coat.J Bacteriol. 2000 Apr;182(7):1828-33. doi: 10.1128/JB.182.7.1828-1833.2000. J Bacteriol. 2000. PMID: 10714986 Free PMC article.
-
A LysM Domain Intervenes in Sequential Protein-Protein and Protein-Peptidoglycan Interactions Important for Spore Coat Assembly in Bacillus subtilis.J Bacteriol. 2019 Jan 28;201(4):e00642-18. doi: 10.1128/JB.00642-18. Print 2019 Feb 15. J Bacteriol. 2019. PMID: 30455281 Free PMC article.
-
SpoVID guides SafA to the spore coat in Bacillus subtilis.J Bacteriol. 2001 May;183(10):3041-9. doi: 10.1128/JB.183.10.3041-3049.2001. J Bacteriol. 2001. PMID: 11325931 Free PMC article.
-
Structure, assembly, and function of the spore surface layers.Annu Rev Microbiol. 2007;61:555-88. doi: 10.1146/annurev.micro.61.080706.093224. Annu Rev Microbiol. 2007. PMID: 18035610 Review.
-
Bacillus subtilis spore coat.Microbiol Mol Biol Rev. 1999 Mar;63(1):1-20. doi: 10.1128/MMBR.63.1.1-20.1999. Microbiol Mol Biol Rev. 1999. PMID: 10066829 Free PMC article. Review.
Cited by
-
The Bacillus subtilis endospore: assembly and functions of the multilayered coat.Nat Rev Microbiol. 2013 Jan;11(1):33-44. doi: 10.1038/nrmicro2921. Epub 2012 Dec 3. Nat Rev Microbiol. 2013. PMID: 23202530 Free PMC article. Review.
-
Ultrastructure of macromolecular assemblies contributing to bacterial spore resistance revealed by in situ cryo-electron tomography.Nat Commun. 2024 Feb 14;15(1):1376. doi: 10.1038/s41467-024-45770-6. Nat Commun. 2024. PMID: 38355696 Free PMC article.
-
CotG-Like Modular Proteins Are Common among Spore-Forming Bacilli.J Bacteriol. 2016 Apr 28;198(10):1513-20. doi: 10.1128/JB.00023-16. Print 2016 May 15. J Bacteriol. 2016. PMID: 26953338 Free PMC article.
-
The coat morphogenetic protein SpoVID is necessary for spore encasement in Bacillus subtilis.Mol Microbiol. 2009 Nov;74(3):634-49. doi: 10.1111/j.1365-2958.2009.06886.x. Epub 2009 Sep 22. Mol Microbiol. 2009. PMID: 19775244 Free PMC article.
-
SpoVID functions as a non-competitive hub that connects the modules for assembly of the inner and outer spore coat layers in Bacillus subtilis.Mol Microbiol. 2018 Nov;110(4):576-595. doi: 10.1111/mmi.14116. Epub 2018 Oct 18. Mol Microbiol. 2018. PMID: 30168214 Free PMC article.
References
-
- Bateman, A., and M. Bycroft. 2000. The structure of a LysM domain from E. coli membrane-bound lytic murein transglycosylase D (MltD). J. Mol. Biol. 299:1113-1119. - PubMed
-
- Birkeland, N. K. 1994. Cloning, molecular characterization, and expression of the genes encoding the lytic functions of lactococcal bacteriophage phi LC3: a dual lysis system of modular design. Can. J. Microbiol. 40:658-665. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

