A novel genetic hierarchy functions during hypaxial myogenesis: Pax3 directly activates Myf5 in muscle progenitor cells in the limb
- PMID: 16951257
- PMCID: PMC1560418
- DOI: 10.1101/gad.382806
A novel genetic hierarchy functions during hypaxial myogenesis: Pax3 directly activates Myf5 in muscle progenitor cells in the limb
Abstract
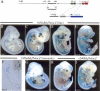
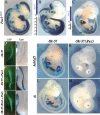
We address the molecular control of myogenesis in progenitor cells derived from the hypaxial somite. Null mutations in Pax3, a key regulator of skeletal muscle formation, lead to cell death in this domain. We have developed a novel allele of Pax3 encoding a Pax3-engrailed fusion protein that acts as a transcriptional repressor. Heterozygote mouse embryos have an attenuated mutant phenotype, with partial conservation of the hypaxial somite and its myogenic derivatives, including some hindlimb muscles. At these sites, expression of Myf5 is compromised, showing that Pax3 acts genetically upstream of this myogenic determination gene. We have characterized a 145-base-pair (bp) regulatory element, at -57.5 kb from Myf5, that directs transgene expression to the mature somite, notably to myogenic cells of the hypaxial domain that form ventral trunk and limb muscles. A Pax3 consensus site in this sequence binds Pax3 in vitro and in vivo. Multimers of the 145-bp sequence direct transgene expression to sites of Pax3 function, and an assay of its activity in the chick embryo shows Pax3 dependence. Mutation of the Pax3 site abolishes all expression controlled by the 145-bp sequence in transgenic mouse embryos. We conclude that Pax3 directly regulates Myf5 in the hypaxial somite and its derivatives.
Figures



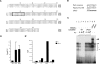
References
-
- Ben-Yair R., Kalcheim C. Lineage analysis of the avian dermomyotome sheet reveals the existence of single cells with both dermal and muscle progenitor fates. Development. 2005;132:689–701. - PubMed
-
- Bladt F., Riethmacher D., Isenmann S., Aguzzi A., Birchmeier C. Essential role for the c-met receptor in the migration of myogenic precursor cells into the limb bud. Nature. 1995;376:768–771. - PubMed
-
- Borycki A.G., Li J., Jin F., Emerson C.P., Epstein J.A. Pax3 functions in cell survival and in pax7 regulation. Development. 1999;126:1665–1674. - PubMed
-
- Braun T., Rudnicki M.A., Arnold H.H., Jaenisch R. Targeted inactivation of the muscle regulatory gene Myf-5 results in abnormal rib development and perinatal death. Cell. 1992;71:369–382. - PubMed
-
- Brohmann H., Jagla K., Birchmeier C. The role of Lbx1 in migration of muscle precursor cells. Development. 2000;127:437–445. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
