Functional characterization of IS1999, an IS4 family element involved in mobilization and expression of beta-lactam resistance genes
- PMID: 16952941
- PMCID: PMC1595497
- DOI: 10.1128/JB.00375-06
Functional characterization of IS1999, an IS4 family element involved in mobilization and expression of beta-lactam resistance genes
Abstract
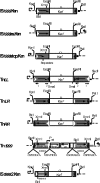
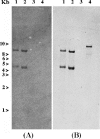
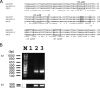
IS1999 and a point mutant derivative, IS1999.2, have been described inserted upstream of emerging antibiotic resistance genes bla(VEB-1) and bla(OXA-48). 5' Rapid amplification of cDNA ends experiments revealed that expression of these beta-lactamase genes was driven by the outward-directed promoter, P(out), located in the IS1999 elements. These findings led us to study IS1999-mediated gene mobilization. Thus, the transposition properties of IS1999 and of IS1999-based composite transposons, made of two copies of IS1999 in different orientations, were investigated. IS1999 or IS1999-based composite transposons were capable of transposing onto the conjugative plasmid pOX38-Gen. Sequence analysis of the insertion sites revealed that IS1999 inserted preferentially into DNA targets containing the consensus sequence NGCNNNGCN. Transposition was more efficient when at least one left inverted repeat end was located at an outside end of the transposon. The transposition frequency of IS1999.2 was 10-fold lower than that of IS1999, and transposition frequencies of the putative natural transposon, Tn1999, were below detection limits of our transposition assay. This reduced transposition frequency of IS1999.2-based elements may result from a lower transcription of the transposase gene, as revealed by reverse transcription-PCR analyses.
Figures


References
-
- Branlant, G., and C. Branlant. 1985. Nucleotide sequence of the Escherichia coli gap gene. Different evolutionary behavior of the NAD+-binding domain and of the catalytic domain of D-glyceraldehyde-3-phosphate dehydrogenase. Eur. J. Biochem. 150:61-66. - PubMed
-
- Casadesus, J., and J. R. Roth. 1989. Transcriptional occlusion of transposon targets. Mol. Gen. Genet. 216:204-209. - PubMed
-
- Chandler, M., M. Clerget, and D. J. Galas. 1982. The transposition frequency of IS1-flanked transposons is a function of their size. J. Mol. Biol. 154:229-243. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources

