Expression and assembly of a functional type IV secretion system elicit extracytoplasmic and cytoplasmic stress responses in Escherichia coli
- PMID: 16952953
- PMCID: PMC1595493
- DOI: 10.1128/JB.00632-06
Expression and assembly of a functional type IV secretion system elicit extracytoplasmic and cytoplasmic stress responses in Escherichia coli
Abstract
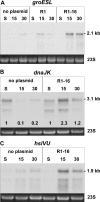
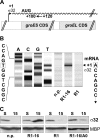
Conditions perturbing protein homeostasis are known to induce cellular stress responses in prokaryotes and eukaryotes. Here we show for the first time that expression and assembly of a functional type IV secretion (T4S) machinery elicit extracytoplasmic and cytoplasmic stress responses in Escherichia coli. After induction of T4S genes by a nutritional upshift and assembly of functional DNA transporters encoded by plasmid R1-16, host cells activated the CpxAR envelope stress signaling system, as revealed by induction or repression of downstream targets of the CpxR response regulator. Furthermore, we observed elevated transcript levels of cytoplasmic stress genes, such as groESL, with a concomitant increase of sigma(32) protein levels in cells expressing T4S genes. A traA null mutant of plasmid R1-16, which lacks the functional gene encoding the major pilus protein pilin, showed distinctly reduced stress responses. These results corroborated our conclusion that the activation of bacterial stress networks was dependent on the presence of functional T4S machinery. Additionally, we detected increased transcription from the rpoHp(1) promoter in the presence of an active T4S system. Stimulation of rpoHp(1) was dependent on the presence of CpxR, suggesting a hitherto undocumented link between CpxAR and sigma(32)-regulated stress networks.
Figures





References
-
- Bardwell, J. C., K. McGovern, and J. Beckwith. 1991. Identification of a protein required for disulfide bond formation in vivo. Cell 67:581-589. - PubMed
-
- Baron, C., D. O'Callaghan, and E. Lanka. 2002. Bacterial secrets of secretion: EuroConference on the biology of type IV secretion processes. Mol. Microbiol. 43:1359-1365. - PubMed
-
- Barrios, A. F., R. Zuo, D. Ren, and T. K. Wood. 2006. Hha, YbaJ, and OmpA regulate Escherichia coli K12 biofilm formation and conjugation plasmids abolish motility. Biotechnol. Bioeng. 93:188-200. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

