Organellar proteomics: turning inventories into insights
- PMID: 16953200
- PMCID: PMC1559674
- DOI: 10.1038/sj.embor.7400780
Organellar proteomics: turning inventories into insights
Abstract
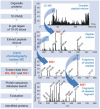
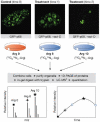
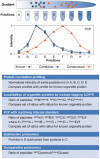
Subcellular organization is yielding to large-scale analysis. Researchers are now applying robust mass-spectrometry-based proteomics methods to obtain an inventory of biochemically isolated organelles that contain hundreds of proteins. High-resolution methods allow accurate protein identification, and novel algorithms can distinguish genuine from co-purifying components. Organellar proteomes have been analysed by bioinformatic methods and integrated with other large-scale data sets. The dynamics of organelles can also be studied by quantitative proteomics, which offers powerful methods that are complementary to fluorescence-based microscopy. Here, we review the emerging trends in this rapidly expanding area and discuss the role of organellar proteomics in the context of functional genomics and systems biology.
Figures



References
-
- Aebersold R, Mann M (2003) Mass spectrometry-based proteomics. Nature 422: 198–207 - PubMed
-
- Andersen JS, Wilkinson CJ, Mayor T, Mortensen P, Nigg EA, Mann M (2003) Proteomic characterization of the human centrosome by protein correlation profiling. Nature 426: 570–574 - PubMed
-
- Andersen JS, Lam YW, Leung AK, Ong SE, Lyon CE, Lamond AI, Mann M (2005) Nucleolar proteome dynamics. Nature 433: 77–83 - PubMed
-
- Broadhead R et al. (2006) Flagellar motility is required for the viability of the bloodstream trypanosome. Nature 440: 224–227 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

