Differentially expressed genes between early and advanced hepatocellular carcinoma (HCC) as a potential tool for selecting liver transplant recipients
- PMID: 16953559
- PMCID: PMC1578766
- DOI: 10.2119/2006-00032.Mas
Differentially expressed genes between early and advanced hepatocellular carcinoma (HCC) as a potential tool for selecting liver transplant recipients
Abstract
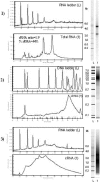
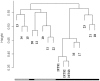
Hepatocellular carcinoma (HCC) is the fifth most common cancer in the world. Liver transplantation (LT) represents a curative treatment for "small" HCC. Preoperative staging is critical in selecting optimal candidates for LT to optimize the use of this scarce resource. From December 1997 to February 2004, 148 patients diagnosed with cirrhosis and HCC were evaluated at our center. After staging, the patients were listed for LT according to United Network for Organ Sharing (UNOS) criteria. When pretransplant liver MRIs were compared with the findings of the explanted livers, 8 of 35 patients (22.8%) were understaged. Three of the 8 patients (37.5%) had recurrence post-LT. A retrospective gene expression profiling study was done using microarray technology for tumor samples in the pretransplant hepatitis C virus (HCV)-HCC understaged patients and in a contemporaneous group of HCV-HCC patients that were accurately staged. Two sample t tests comparing the early versus advanced HCV-HCCs with respect to gene expression showed an important set of genes differentially expressed among the samples. Hierarchical clustering analysis of the gene expression profiling classified 93.8% of the total tumor samples and 85.7% of the understaged samples in concordance with the explanted pathological staging. We found a distinctive pattern of gene expression between early and advanced HCV-HCCs. These results suggest that gene expression profiling could improve the pre-LT HCC staging to more closely mimic the explant pathology. Whether gene expression profiling of HCC will be refined to the point of predicting potential metastatic biologic behavior to predict post-LT recurrence will require longitudinal prospective study of this gene array technology.
Figures
Similar articles
-
Genes associated with progression and recurrence of hepatocellular carcinoma in hepatitis C patients waiting and undergoing liver transplantation: preliminary results.Transplantation. 2007 Apr 15;83(7):973-81. doi: 10.1097/01.tp.0000258643.05294.0b. Transplantation. 2007. PMID: 17460570
-
Liver transplantation recipients with nonalcoholic steatohepatitis have lower risk hepatocellular carcinoma.Liver Transpl. 2017 Aug;23(8):1015-1022. doi: 10.1002/lt.24764. Epub 2017 Jun 26. Liver Transpl. 2017. PMID: 28340509
-
Hepatocellular carcinoma in HCV-infected patients awaiting liver transplantation: genes involved in tumor progression.Liver Transpl. 2004 May;10(5):607-20. doi: 10.1002/lt.20118. Liver Transpl. 2004. PMID: 15108252
-
LI-RADS and transplantation for hepatocellular carcinoma.Abdom Radiol (NY). 2018 Jan;43(1):193-202. doi: 10.1007/s00261-017-1210-8. Abdom Radiol (NY). 2018. PMID: 28612162 Review.
-
Molecular profiling of hepatocellular carcinomas by cDNA microarray.World J Gastroenterol. 2005 Jan 28;11(4):463-8. doi: 10.3748/wjg.v11.i4.463. World J Gastroenterol. 2005. PMID: 15641127 Free PMC article. Review.
Cited by
-
Identifying genes progressively silenced in preneoplastic and neoplastic liver tissues.Int J Comput Biol Drug Des. 2010;3(1):52-67. doi: 10.1504/IJCBDD.2010.034499. Epub 2010 Aug 5. Int J Comput Biol Drug Des. 2010. PMID: 20693610 Free PMC article.
-
An integrative meta-analysis of microRNAs in hepatocellular carcinoma.Genomics Proteomics Bioinformatics. 2013 Dec;11(6):354-67. doi: 10.1016/j.gpb.2013.05.007. Epub 2013 Nov 25. Genomics Proteomics Bioinformatics. 2013. PMID: 24287119 Free PMC article.
-
Genes involved in viral carcinogenesis and tumor initiation in hepatitis C virus-induced hepatocellular carcinoma.Mol Med. 2009 Mar-Apr;15(3-4):85-94. doi: 10.2119/molmed.2008.00110. Epub 2008 Dec 15. Mol Med. 2009. PMID: 19098997 Free PMC article.
-
Functional and topological properties in hepatocellular carcinoma transcriptome.PLoS One. 2012;7(4):e35510. doi: 10.1371/journal.pone.0035510. Epub 2012 Apr 23. PLoS One. 2012. PMID: 22539975 Free PMC article.
-
Transcriptomic profiles of tumor-associated neutrophils reveal prominent roles in enhancing angiogenesis in liver tumorigenesis in zebrafish.Sci Rep. 2019 Feb 6;9(1):1509. doi: 10.1038/s41598-018-36605-8. Sci Rep. 2019. PMID: 30728369 Free PMC article.
References
-
- Parkin DM, Bray F, Ferlay J, Pisani P. Estimating the world cancer burden: Globocan 2000. Int J Cancer. 2001;94:153–6. - PubMed
-
- El-Serag HB, Mason AC. Rising incidence of hepatocellular carcinoma in the United States. N Engl J Med. 1999;340:745–50. - PubMed
-
- Daniele B, Bencivenga A, Megna AS, Tinessa V. Alpha-fetoprotein and ultrasonography screening for hepatocellular carcinoma. Gastroenterology. 2004;12:S108–12. - PubMed
-
- Soresi M, Magliarisi C, Campagna P, et al. Usefulness of alpha-fetoprotein in the diagnosis of hepatocellular carcinoma. Anticancer Res. 2003;23:1747–53. - PubMed
-
- Di Bisceglie AM. Issues in screening and surveillance for hepatocellular carcinoma. Gastroenterology. 2004;127:S104–7. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
