Genetic analysis of Thailand hantavirus in Bandicota indica trapped in Thailand
- PMID: 16953877
- PMCID: PMC1578555
- DOI: 10.1186/1743-422X-3-72
Genetic analysis of Thailand hantavirus in Bandicota indica trapped in Thailand
Abstract
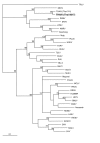
Sixty one tissue samples from several rodent species trapped in five provinces of Thailand were examined for the presence of hantaviral markers by enzyme-immunoassay and immunoblotting. Four samples, all from the great bandicoot rat Bandicota indica, were confirmed positive for the hantaviral N-antigen. Two of them were trapped in Nakhon Pathom province, the other two in Nakhon Ratchasima province, approximately 250 km from the other trapping site. When analysed by RT-nested PCR, all four rodents were found positive for the hantaviral S- and M-segment nucleotide sequences. Genetic analysis revealed that the four newly described wild-type strains belong to Thailand hantavirus. On the phylogenetic trees they formed a well-supported cluster within the group of Murinae-associated hantaviruses and shared a recent common ancestor with Seoul virus.
Figures


References
-
- Nichol ST, Beaty BJ, Elliott RM, Goldbach R, Plyusnin A, Schmaljohn CS, Tesh RB. Bunyaviridae. In: Fauquet CM, Mayo MA, Maniloff J, Desselberger U, Ball LA, editor. Virus taxonomy VIIIth report of the International Committee on Taxonomy of Viruses. Amsterdam: Elsevier Academic Press; 2005. pp. 695–716.
-
- Lundkvist Å, Plyusnin A. Molecular epidemiology of hantavirus infections. In: Leitner T, editor. The Molecular Epidemiology of Human Viruses. Kluwer Academic Publishers; 2002. pp. 351–384.
-
- Plyusnin A, Morzunov S. Virus evolution and genetic diversity of hantaviruses and their rodent hosts. In: Schmaljohn C, Nichol SN, editor. Curr Top Microbiol Immunol. Vol. 256. 2001. pp. 47–75. - PubMed
-
- Nemirov K, Vaheri A, Plyusnin A. Hantaviruses:co-evolution with natural hosts. Recent Res Devel Virol. 2004;6:201–228.
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

