Differential detection of Entamoeba histolytica, Entamoeba dispar, and Entamoeba moshkovskii by a single-round PCR assay
- PMID: 16954247
- PMCID: PMC1594701
- DOI: 10.1128/JCM.00778-06
Differential detection of Entamoeba histolytica, Entamoeba dispar, and Entamoeba moshkovskii by a single-round PCR assay
Abstract
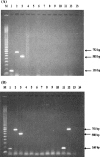
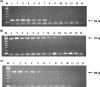
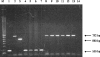
A single-round PCR assay was developed for detection and differential diagnosis of the three Entamoeba species found in humans, Entamoeba moshkovskii, Entamoeba histolytica, and Entamoeba dispar, that are morphologically identical as both cysts and trophozoites. A conserved forward primer was derived from the middle of the small-subunit rRNA gene, and reverse primers were designed from signature sequences specific to each of these three Entamoeba species. PCR generates a 166-bp product with E. histolytica DNA, a 752-bp product with E. dispar DNA, and a 580-bp product with E. moshkovskii DNA. Thirty clinical specimens were examined, and the species present were successfully detected and differentiated using this assay. It was possible to detect as little as 10 pg of E. moshkovskii and E. histolytica DNA, while for E. dispar the sensitivity was about 20 pg of DNA. Testing with DNA from different pathogens, including bacteria and other protozoa, confirmed the high specificity of the assay. We propose the use of this PCR assay as an accurate, rapid, and effective diagnostic method for the detection and discrimination of these three morphologically indistinguishable Entamoeba species in both routine diagnosis of amoebiasis and epidemiological surveys.
Figures
References
-
- Acuna-Soto, R., J. Samuelson, P. De-Girolami, et al. 1994. Application of the polymerase chain reaction to the epidemiology of pathogenic and nonpathogenic Entamoeba histolytica. Am. J. Trop. Med. Hyg. 48:58-70. - PubMed
-
- Clark, C. G., and L. S. Diamond. 1991. The Laredo strain and other Entamoeba histolytica-like amoebae are Entamoeba moshkovskii. Mol. Biochem. Parasitol. 46:11-18. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

