Differential impact of Ink4a and Arf on hematopoietic stem cells and their bone marrow microenvironment in Bmi1-deficient mice
- PMID: 16954369
- PMCID: PMC2118102
- DOI: 10.1084/jem.20052477
Differential impact of Ink4a and Arf on hematopoietic stem cells and their bone marrow microenvironment in Bmi1-deficient mice
Abstract
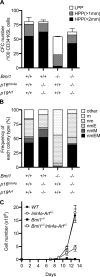
The polycomb group (PcG) protein Bmi1 plays an essential role in the self-renewal of hematopoietic and neural stem cells. Derepression of the Ink4a/Arf gene locus has been largely attributed to Bmi1-deficient phenotypes in the nervous system. However, its role in hematopoietic stem cell (HSC) self-renewal remained undetermined. In this study, we show that derepressed p16(Ink4a) and p19(Arf) in Bmi1-deficient mice were tightly associated with a loss of self-renewing HSCs. The deletion of both Ink4a and Arf genes substantially restored the self-renewal capacity of Bmi1(-/-) HSCs. Thus, Bmi1 regulates HSCs by acting as a critical failsafe against the p16(Ink4a)- and p19(Arf)-dependent premature loss of HSCs. We further identified a novel role for Bmi1 in the organization of a functional bone marrow (BM) microenvironment. The BM microenvironment in Bmi1(-/-) mice appeared severely defective in supporting hematopoiesis. The deletion of both Ink4a and Arf genes did not considerably restore the impaired BM microenvironment, leading to a sustained postnatal HSC depletion in Bmi1(-/-)Ink4a-Arf(-/-) mice. Our findings unveil a differential role of derepressed Ink4a and Arf on HSCs and their BM microenvironment in Bmi1-deficient mice. Collectively, Bmi1 regulates self-renewing HSCs in both cell-autonomous and nonautonomous manners.
Figures



References
-
- Lund, A.H., and M. van Lohuizen. 2004. Polycomb complexes and silencing mechanisms. Curr. Opin. Cell Biol. 16:239–246. - PubMed
-
- Valk-Lingbeek, M.E., S.W.M. Bruggeman, and M. van Lohuizen. 2004. Stem cells and cancer: the polycomb connection. Cell. 118:409–418. - PubMed
-
- Iwama, A., H. Oguro, M. Negishi, Y. Kato, and H. Nakauchi. 2005. Epigenetic regulation of hematopoietic stem cell self-renewal by polycomb group genes. Int. J. Hematol. 81:294–300. - PubMed
-
- van der Lugt, N.M., J. Domen, K. Linders, M. van Roon, E. Robanus-Maandag, H. te Riele, M. van der Valk, J. Deschamps, M. Sofroniew, M. van Lohuizen, and A. Berns. 1994. Posterior transformation, neurological abnormalities, and severe hematopoietic defects in mice with a targeted deletion of the bmi-1 proto-oncogene. Genes Dev. 8:757–769. - PubMed
-
- Lessard, J., and G. Sauvageau. 2003. Bmi-1 determines proliferative capacity of normal and leukemic stem cells. Nature. 423:255–260. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

