Cotranscriptional recruitment to the mRNA export receptor Mex67p contributes to nuclear pore anchoring of activated genes
- PMID: 16954382
- PMCID: PMC1636739
- DOI: 10.1128/MCB.00870-06
Cotranscriptional recruitment to the mRNA export receptor Mex67p contributes to nuclear pore anchoring of activated genes
Abstract
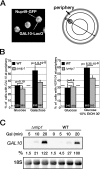
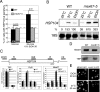
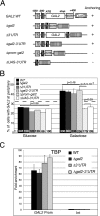
Transcription activation of some Saccharomyces cerevisiae genes is paralleled by their repositioning to the nuclear periphery, but the mechanism underlying gene anchoring is poorly defined. We show that the nuclear pore complex-associated Mlp1p and the shuttling mRNA export receptor Mex67p contribute to the stable association of the activated GAL10 and HSP104 genes with the nuclear periphery. However, we find no obligatory link between gene positioning and gene expression. Furthermore, gene anchoring correlates with the cotranscriptional recruitment of Mex67p to transcribing genes. Notably, the association of Mex67p with chromatin is not mediated by RNA. Interestingly, a mutant GAL2 gene lacking the coding region is still able to recruit Mex67p upon transcriptional activation and to relocate to the nuclear periphery. Together these data suggest that, at least for GAL2, nascent messenger ribonucleoprotein does not play a major role in gene anchoring and that the early recruitment of Mex67p contributes to gene repositioning by virtue of an RNA-independent process.
Figures



References
-
- Andrulis, E. D., A. M. Neiman, D. C. Zappulla, and R. Sternglanz. 1998. Perinuclear localization of chromatin facilitates transcriptional silencing. Nature 394:592-595. - PubMed
-
- Cabal, G. G., A. Genovesio, S. Rodriguez-Navarro, C. Zimmer, O. Gadal, A. Lesne, H. Buc, F. Feuerbach-Fournier, J. C. Olivo-Marin, E. C. Hurt, and U. Nehrbass. 2006. SAGA interacting factors confine subdiffusion of transcribed genes to the nuclear envelope. Nature 441:770-773. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
