Many novel mammalian microRNA candidates identified by extensive cloning and RAKE analysis
- PMID: 16954537
- PMCID: PMC1581438
- DOI: 10.1101/gr.5159906
Many novel mammalian microRNA candidates identified by extensive cloning and RAKE analysis
Abstract
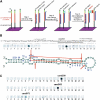
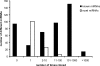
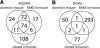
MicroRNAs are 20- to 23-nucleotide RNA molecules that can regulate gene expression. Currently > 400 microRNAs have been experimentally identified in mammalian genomes, whereas estimates go up to 1000 and beyond. Here we show that many more mammalian microRNAs exist. We discovered novel microRNA candidates using two approaches: testing of computationally predicted microRNAs by a modified microarray-based detection system, and cloning and sequencing of large numbers of small RNAs from different human and mouse tissues. Together these efforts experimentally identified 348 novel mouse and 81 novel human microRNA candidate genes. Most novel microRNAs candidates are not conserved beyond mammals, and ~10% are taxon-specific. Our analyses indicate that the entire microRNA repertoire is not remotely exhausted.
Figures

References
-
- Altuvia Y., Landgraf P., Lithwick G., Elefant N., Pfeffer S., Aravin A., Brownstein M.J., Tuschl T., Margalit H., Landgraf P., Lithwick G., Elefant N., Pfeffer S., Aravin A., Brownstein M.J., Tuschl T., Margalit H., Lithwick G., Elefant N., Pfeffer S., Aravin A., Brownstein M.J., Tuschl T., Margalit H., Elefant N., Pfeffer S., Aravin A., Brownstein M.J., Tuschl T., Margalit H., Pfeffer S., Aravin A., Brownstein M.J., Tuschl T., Margalit H., Aravin A., Brownstein M.J., Tuschl T., Margalit H., Brownstein M.J., Tuschl T., Margalit H., Tuschl T., Margalit H., Margalit H. Clustering and conservation patterns of human microRNAs. Nucleic Acids Res. 2005;33:2697–2706. - PMC - PubMed
-
- Alvarez-Garcia I., Miska E.A., Miska E.A. MicroRNA functions in animal development and human disease. Development. 2005;132:4653–4662. - PubMed
-
- Ambros V., Bartel B., Bartel D.P., Burge C.B., Carrington J.C., Chen X., Dreyfuss G., Eddy S.R., Griffiths-Jones S., Marshall M., Bartel B., Bartel D.P., Burge C.B., Carrington J.C., Chen X., Dreyfuss G., Eddy S.R., Griffiths-Jones S., Marshall M., Bartel D.P., Burge C.B., Carrington J.C., Chen X., Dreyfuss G., Eddy S.R., Griffiths-Jones S., Marshall M., Burge C.B., Carrington J.C., Chen X., Dreyfuss G., Eddy S.R., Griffiths-Jones S., Marshall M., Carrington J.C., Chen X., Dreyfuss G., Eddy S.R., Griffiths-Jones S., Marshall M., Chen X., Dreyfuss G., Eddy S.R., Griffiths-Jones S., Marshall M., Dreyfuss G., Eddy S.R., Griffiths-Jones S., Marshall M., Eddy S.R., Griffiths-Jones S., Marshall M., Griffiths-Jones S., Marshall M., Marshall M., et al. A uniform system for microRNA annotation. RNA. 2003;9:277–279. - PMC - PubMed
-
- Aravin A., Tuschl T., Tuschl T. Identification and characterization of small RNAs involved in RNA silencing. Bartel, D.P. 2004. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 116: 281–297. FEBS Lett. 2005;579:5830–5840. - PubMed
-
- Bentwich I., Avniel A., Karov Y., Aharonov R., Gilad S., Barad O., Barzilai A., Einat P., Einav U., Meiri E., Avniel A., Karov Y., Aharonov R., Gilad S., Barad O., Barzilai A., Einat P., Einav U., Meiri E., Karov Y., Aharonov R., Gilad S., Barad O., Barzilai A., Einat P., Einav U., Meiri E., Aharonov R., Gilad S., Barad O., Barzilai A., Einat P., Einav U., Meiri E., Gilad S., Barad O., Barzilai A., Einat P., Einav U., Meiri E., Barad O., Barzilai A., Einat P., Einav U., Meiri E., Barzilai A., Einat P., Einav U., Meiri E., Einat P., Einav U., Meiri E., Einav U., Meiri E., Meiri E., et al. Identification of hundreds of conserved and nonconserved human microRNAs. Nat. Genet. 2005;37:766–770. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
