MicroRNAs and other small RNAs enriched in the Arabidopsis RNA-dependent RNA polymerase-2 mutant
- PMID: 16954541
- PMCID: PMC1581437
- DOI: 10.1101/gr.5530106
MicroRNAs and other small RNAs enriched in the Arabidopsis RNA-dependent RNA polymerase-2 mutant
Abstract
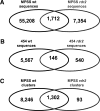
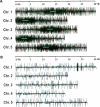
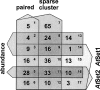
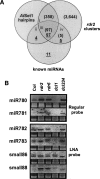
The Arabidopsis genome contains a highly complex and abundant population of small RNAs, and many of the endogenous siRNAs are dependent on RNA-Dependent RNA Polymerase 2 (RDR2) for their biogenesis. By analyzing an rdr2 loss-of-function mutant using two different parallel sequencing technologies, MPSS and 454, we characterized the complement of miRNAs expressed in Arabidopsis inflorescence to considerable depth. Nearly all known miRNAs were enriched in this mutant and we identified 13 new miRNAs, all of which were relatively low abundance and constitute new families. Trans-acting siRNAs (ta-siRNAs) were even more highly enriched. Computational and gel blot analyses suggested that the minimal number of miRNAs in Arabidopsis is approximately 155. The size profile of small RNAs in rdr2 reflected enrichment of 21-nt miRNAs and other classes of siRNAs like ta-siRNAs, and a significant reduction in 24-nt heterochromatic siRNAs. Other classes of small RNAs were found to be RDR2-independent, particularly those derived from long inverted repeats and a subset of tandem repeats. The small RNA populations in other Arabidopsis small RNA biogenesis mutants were also examined; a dcl2/3/4 triple mutant showed a similar pattern to rdr2, whereas dcl1-7 and rdr6 showed reductions in miRNAs and ta-siRNAs consistent with their activities in the biogenesis of these types of small RNAs. Deep sequencing of mutants provides a genetic approach for the dissection and characterization of diverse small RNA populations and the identification of low abundance miRNAs.
Figures


References
-
- Adai A., Johnson C., Mlotshwa S., Archer-Evans S., Manocha V., Vance V., Sundaresan V., Johnson C., Mlotshwa S., Archer-Evans S., Manocha V., Vance V., Sundaresan V., Mlotshwa S., Archer-Evans S., Manocha V., Vance V., Sundaresan V., Archer-Evans S., Manocha V., Vance V., Sundaresan V., Manocha V., Vance V., Sundaresan V., Vance V., Sundaresan V., Sundaresan V. Computational prediction of miRNAs in Arabidopsis thaliana . Genome Res. 2005;15:78–91. - PMC - PubMed
-
- Allen E., Xie Z., Gustafson A.M., Sung G.H., Spatafora J.W., Carrington J.C., Xie Z., Gustafson A.M., Sung G.H., Spatafora J.W., Carrington J.C., Gustafson A.M., Sung G.H., Spatafora J.W., Carrington J.C., Sung G.H., Spatafora J.W., Carrington J.C., Spatafora J.W., Carrington J.C., Carrington J.C. Evolution of microRNA genes by inverted duplication of target gene sequences in Arabidopsis thaliana . Nat. Genet. 2004;36:1282–1290. - PubMed
-
- Allen E., Xie Z., Gustafson A.M., Carrington J.C., Xie Z., Gustafson A.M., Carrington J.C., Gustafson A.M., Carrington J.C., Carrington J.C. microRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell. 2005;121:207–221. - PubMed
-
- Ambros V., Bartel B., Bartel D.P., Burge C.B., Carrington J.C., Chen X., Dreyfuss G., Eddy S.R., Griffiths-Jones S., Marshall M., Bartel B., Bartel D.P., Burge C.B., Carrington J.C., Chen X., Dreyfuss G., Eddy S.R., Griffiths-Jones S., Marshall M., Bartel D.P., Burge C.B., Carrington J.C., Chen X., Dreyfuss G., Eddy S.R., Griffiths-Jones S., Marshall M., Burge C.B., Carrington J.C., Chen X., Dreyfuss G., Eddy S.R., Griffiths-Jones S., Marshall M., Carrington J.C., Chen X., Dreyfuss G., Eddy S.R., Griffiths-Jones S., Marshall M., Chen X., Dreyfuss G., Eddy S.R., Griffiths-Jones S., Marshall M., Dreyfuss G., Eddy S.R., Griffiths-Jones S., Marshall M., Eddy S.R., Griffiths-Jones S., Marshall M., Griffiths-Jones S., Marshall M., Marshall M., et al. A uniform system for microRNA annotation. RNA. 2003;9:277–279. - PMC - PubMed
-
- Arazi T., Talmor-Neiman M., Stav R., Riese M., Huijser P., Baulcombe D.C., Talmor-Neiman M., Stav R., Riese M., Huijser P., Baulcombe D.C., Stav R., Riese M., Huijser P., Baulcombe D.C., Riese M., Huijser P., Baulcombe D.C., Huijser P., Baulcombe D.C., Baulcombe D.C. Cloning and characterization of micro-RNAs from moss. Plant J. 2005;43:837–848. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
