Recombination and selection in the evolution of picornaviruses and other Mammalian positive-stranded RNA viruses
- PMID: 16956935
- PMCID: PMC1642140
- DOI: 10.1128/JVI.01076-06
Recombination and selection in the evolution of picornaviruses and other Mammalian positive-stranded RNA viruses
Abstract
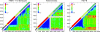
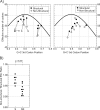
Picornaviridae are a large virus family causing widespread, often pathogenic infections in humans and other mammals. Picornaviruses are genetically and antigenically highly diverse, with evidence for complex evolutionary histories in which recombination plays a major part. To investigate the nature of recombination and selection processes underlying the evolution of serotypes within different picornavirus genera, large-scale analysis of recombination frequencies and sites, segregation by serotype within each genus, and sequence selection and composition was performed, and results were compared with those for other nonenveloped positive-stranded viruses (astroviruses and human noroviruses) and with flavivirus and alphavirus control groups. Enteroviruses, aphthoviruses, and teschoviruses showed phylogenetic segregation by serotype only in the structural region; lack of segregation elsewhere was attributable to extensive interserotype recombination. Nonsegregating viruses also showed several characteristic sequence divergence and composition differences between genome regions that were absent from segregating virus control groups, such as much greater amino acid sequence divergence in the structural region, markedly elevated ratios of nonsynonymous-to-synonymous substitutions, and differences in codon usage. These properties were shared with other picornavirus genera, such as the parechoviruses and erboviruses. The nonenveloped astroviruses and noroviruses similarly showed high frequencies of recombination, evidence for positive selection, and differential codon use in the capsid region, implying similar underlying evolutionary mechanisms and pressures driving serotype differentiation. This process was distinct from more-recent sequence evolution generating diversity within picornavirus serotypes, in which neutral or purifying selection was prominent. Overall, this study identifies common themes in the diversification process generating picornavirus serotypes that contribute to understanding of their evolution and pathogenicity.
Figures





References
-
- Andersson, P., K. Edman, and A. M. Lindberg. 2002. Molecular analysis of the echovirus 18 prototype: evidence of interserotypic recombination with echovirus 9. Virus Res. 85:71-83. - PubMed
-
- Ando, T., J. S. Noel, and R. L. Fankhauser. 2000. Genetic classification of “Norwalk-like viruses.” J. Infect. Dis. 181(Suppl. 2):S336-S348. - PubMed
-
- Baranowski, E., C. M. Ruiz-Jarabo, and E. Domingo. 2001. Evolution of cell recognition by viruses. Science 292:1102-1105. - PubMed
-
- Belliot, G., H. Laveran, and S. S. Monroe. 1997. Detection and genetic differentiation of human astroviruses: phylogenetic grouping varies by coding region. Arch. Virol. 142:1323-1334. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

