Cooperative involvement of the S1 and S2 subunits of the murine coronavirus spike protein in receptor binding and extended host range
- PMID: 16956938
- PMCID: PMC1642182
- DOI: 10.1128/JVI.00950-06
Cooperative involvement of the S1 and S2 subunits of the murine coronavirus spike protein in receptor binding and extended host range
Abstract
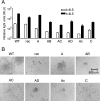
To study the process of spike (S)-receptor interaction during coronavirus entry, we evaluated the contributions of mutations in different regions of the murine hepatitis virus (MHV) S protein to natural receptor murine carcinoembryonic antigen-related cell adhesion molecule 1a (CEACAM1a) dependence and to the acquisition of extended host range. Extended-host-range variants of MHV strain A59 were previously obtained from persistently infected cells (J. H. Schickli, B. D. Zelus, D. E. Wentworth, S. G. Sawicki, and K. V. Holmes, J. Virol. 71:9499-9504, 1997). These variant viruses contain several mutations in the S protein that confer to the viruses the ability to enter cells in a heparan sulfate-dependent manner (C. A. de Haan, Z. Li, E. te Lintelo, B. J. Bosch, B. J. Haijema, and P. J. M. Rottier, J. Virol. 79:14451-14456, 2005). While the parental MHV-A59 is fully dependent on murine CEACAM1a for its entry, viruses carrying the variant mutations in the amino-terminal part of their S protein had become dependent on both CEACAM1a and heparan sulfate. Substitutions in a restricted, downstream part of the S protein encompassing heptad repeat region 1 (HR1) and putative fusion peptide (FP) did not alter the CEACAM1a dependence. However, when the mutations in both parts of the S protein were combined, the resulting viruses became independent of CEACAM1a and acquired the extended host range. In addition, these viruses showed a decreased binding to and inhibition by soluble CEACAM1a. The observations suggest that the amino-terminal region of the S protein, including the receptor-binding domain, and a region in the central part of the S protein containing HR1 and FP, i.e., regions far apart in the linear sequence, communicate and may even interact physically in the higher-order structure of the spike.
Figures


Similar articles
-
The N-terminal region of the murine coronavirus spike glycoprotein is associated with the extended host range of viruses from persistently infected murine cells.J Virol. 2004 Sep;78(17):9073-83. doi: 10.1128/JVI.78.17.9073-9083.2004. J Virol. 2004. PMID: 15308703 Free PMC article.
-
Amino acid substitutions in the S2 subunit of mouse hepatitis virus variant V51 encode determinants of host range expansion.J Virol. 2008 Feb;82(3):1414-24. doi: 10.1128/JVI.01674-07. Epub 2007 Nov 21. J Virol. 2008. PMID: 18032498 Free PMC article.
-
Amino acid substitutions and an insertion in the spike glycoprotein extend the host range of the murine coronavirus MHV-A59.Virology. 2004 Jul 1;324(2):510-24. doi: 10.1016/j.virol.2004.04.005. Virology. 2004. PMID: 15207636 Free PMC article.
-
Structure, Function, and Evolution of Coronavirus Spike Proteins.Annu Rev Virol. 2016 Sep 29;3(1):237-261. doi: 10.1146/annurev-virology-110615-042301. Epub 2016 Aug 25. Annu Rev Virol. 2016. PMID: 27578435 Free PMC article. Review.
-
Pathogenesis of murine coronavirus in the central nervous system.J Neuroimmune Pharmacol. 2010 Sep;5(3):336-54. doi: 10.1007/s11481-010-9202-2. Epub 2010 Apr 6. J Neuroimmune Pharmacol. 2010. PMID: 20369302 Free PMC article. Review.
Cited by
-
Membrane Protein of Human Coronavirus NL63 Is Responsible for Interaction with the Adhesion Receptor.J Virol. 2019 Sep 12;93(19):e00355-19. doi: 10.1128/JVI.00355-19. Print 2019 Oct 1. J Virol. 2019. PMID: 31315999 Free PMC article.
-
Coronavirus Spike Protein and Tropism Changes.Adv Virus Res. 2016;96:29-57. doi: 10.1016/bs.aivir.2016.08.004. Epub 2016 Sep 13. Adv Virus Res. 2016. PMID: 27712627 Free PMC article. Review.
-
Reduction of Cell Fusion by Deletion in the Hypervariable Region of the Spike Protein of Mouse Hepatitis Virus.Viruses. 2022 Feb 15;14(2):398. doi: 10.3390/v14020398. Viruses. 2022. PMID: 35215991 Free PMC article.
-
Contributions of the S2 spike ectodomain to attachment and host range of infectious bronchitis virus.Virus Res. 2013 Nov 6;177(2):127-37. doi: 10.1016/j.virusres.2013.09.006. Epub 2013 Sep 13. Virus Res. 2013. PMID: 24041648 Free PMC article.
-
Non-invasive imaging of mouse hepatitis coronavirus infection reveals determinants of viral replication and spread in vivo.Cell Microbiol. 2009 May;11(5):825-41. doi: 10.1111/j.1462-5822.2009.01298.x. Epub 2009 Feb 10. Cell Microbiol. 2009. PMID: 19215224 Free PMC article.
References
-
- Bosch, B. J., B. E. Martina, R. Van Der Zee, J. Lepault, B. J. Haijema, C. Versluis, A. J. Heck, R. De Groot, A. D. Osterhaus, and P. J. Rottier. 2004. Severe acute respiratory syndrome coronavirus (SARS-CoV) infection inhibition using spike protein heptad repeat-derived peptides. Proc. Natl. Acad. Sci. USA 101:8455-8460. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

