In vitro resistance to the human immunodeficiency virus type 1 maturation inhibitor PA-457 (Bevirimat)
- PMID: 16956950
- PMCID: PMC1642185
- DOI: 10.1128/JVI.01369-06
In vitro resistance to the human immunodeficiency virus type 1 maturation inhibitor PA-457 (Bevirimat)
Abstract
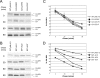
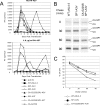
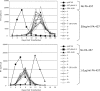
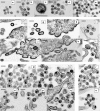
3-O-(3',3'-dimethylsuccinyl)betulinic acid (PA-457 or bevirimat) potently inhibits human immunodeficiency virus type 1 (HIV-1) maturation by blocking a late step in the Gag processing pathway, specifically the cleavage of SP1 from the C terminus of capsid (CA). To gain insights into the mechanism(s) by which HIV-1 could evolve resistance to PA-457 and to evaluate the likelihood of such resistance arising in PA-457-treated patients, we sought to identify and characterize a broad spectrum of HIV-1 variants capable of conferring resistance to this compound. Numerous independent rounds of selection repeatedly identified six single-amino-acid substitutions that independently confer PA-457 resistance: three at or near the C terminus of CA (CA-H226Y, -L231F, and -L231M) and three at the first and third residues of SP1 (SP1-A1V, -A3T, and -A3V). We determined that mutations CA-H226Y, CA-L231F, CA-L231M, and SP1-A1V do not impose a significant replication defect on HIV-1 in culture. In contrast, mutations SP1-A3V and -A3T severely impaired virus replication and inhibited virion core condensation. The replication defect imposed by SP1-A3V was reversed by a second-site compensatory mutation in CA (CA-G225S). Intriguingly, high concentrations of PA-457 enhanced the maturation of SP1 residue 3 mutants. The different phenotypes associated with mutations that confer PA-457 resistance suggest the existence of multiple mechanisms by which HIV-1 can evolve resistance to this maturation inhibitor. These findings have implications for the ongoing development of PA-457 to treat HIV-1 infection in vivo.
Figures







References
-
- Abdurahman, S., S. Hoglund, L. Goobar-Larsson, and A. Vahlne. 2004. Selected amino acid substitutions in the C-terminal region of human immunodeficiency virus type 1 capsid protein affect virus assembly and release. J Gen. Virol. 85:2903-2913. - PubMed
-
- Adamson, C. S., and I. M. Jones. 2004. The molecular basis of HIV capsid assembly—five years of progress. Rev. Med. Virol. 14:107-121. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

