Crystal structure of human histone lysine-specific demethylase 1 (LSD1)
- PMID: 16956976
- PMCID: PMC1599895
- DOI: 10.1073/pnas.0606381103
Crystal structure of human histone lysine-specific demethylase 1 (LSD1)
Abstract
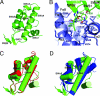
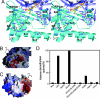
Lysine-specific demethylase 1 (LSD1) was recently identified as the first histone demethylase that specifically demethylates monomethylated and dimethylated histone H3 at K4. It is a component of the CoREST and other corepressor complexes and plays an important role in silencing neuronal-specific genes in nonneuronal cells, but the molecular mechanisms of its action remain unclear. The 2.8-A-resolution crystal structure of the human LSD1 reveals that LSD1 defines a new subfamily of FAD-dependent oxidases. The active center of LSD1 is characterized by a remarkable 1,245-A3 substrate-binding cavity with a highly negative electrostatic potential. Although the protein core of LSD1 resembles other flavoenzymes, its enzymatic activity and functions require two additional structural modules: an N-terminal SWIRM domain important for protein stability and a large insertion in the catalytic domain indispensable both for the demethylase activity and the interaction with CoREST. These results provide a framework for further probing the catalytic mechanism and the functional roles of LSD1.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Fischle W, Wang Y, Allis CD. Curr Opin Cell Biol. 2003;15:172–183. - PubMed
-
- Margueron R, Trojer P, Reinberg D. Curr Opin Genet Dev. 2005;15:163–176. - PubMed
-
- Martin C, Zhang Y. Nat Rev Mol Cell Biol. 2005;6:838–849. - PubMed
-
- Shi Y, Lan F, Matson C, Mulligan P, Whetstine JR, Cole PA, Casero RA, Shi Y. Cell. 2004;119:941–953. - PubMed
-
- Tsukada Y, Fang J, Erdjument-Bromage H, Warren ME, Borchers CH, Tempst P, Zhang Y. Nature. 2006;439:811–816. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

