Genotyping hepatitis C viruses from Southeast Asia by a novel line probe assay that simultaneously detects core and 5' untranslated regions
- PMID: 16957039
- PMCID: PMC1698374
- DOI: 10.1128/JCM.01122-06
Genotyping hepatitis C viruses from Southeast Asia by a novel line probe assay that simultaneously detects core and 5' untranslated regions
Abstract
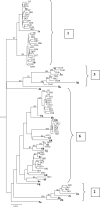
Hepatitis C viruses (HCVs) display a high level of sequence diversity and are currently divided into six genotypes. A line probe assay (LiPA), which targets the 5' untranslated region (5'UTR) of the HCV genome, is widely used for genotyping. However, this assay cannot distinguish many genotype 6 subtypes from genotype 1 due to high sequence similarity in the 5'UTR. We investigated the accuracy of a new generation LiPA (VERSANT HCV genotype 2.0 assay), in which genotyping is based on 5'UTR and core sequences, by testing 75 selected HCV RNA-positive sera from Southeast Asia (Vietnam and Thailand). For comparison, sera were tested on the 5'UTR based VERSANT HCV genotype assay and processed for sequence analysis of the 5'UTR-to-core and NS5b regions as well. Phylogenetic analysis of both regions revealed the presence of genotype 1, 2, 3, and 6 viruses. Using the new LiPA assay, genotypes 6c to 6l and 1a/b samples were more accurately genotyped than with the previous test only targeting the 5'UTR (96% versus 71%, respectively). These results indicate that the VERSANT HCV genotype 2.0 assay is able to discriminate genotypes 6c to 6l from genotype 1 and allows a more accurate identification of genotype 1a from 1b by using the genotype-specific core information.
Figures
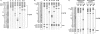

Similar articles
-
Improvement of hepatitis C virus (HCV) genotype determination with the new version of the INNO-LiPA HCV assay.J Clin Microbiol. 2007 Apr;45(4):1140-5. doi: 10.1128/JCM.01982-06. Epub 2007 Jan 24. J Clin Microbiol. 2007. PMID: 17251399 Free PMC article.
-
Evaluation of a new-generation line-probe assay that detects 5' untranslated and core regions to genotype and subtype hepatitis C virus.Am J Clin Pathol. 2007 Aug;128(2):300-4. doi: 10.1309/RBAAC0HJFWRJB1HD. Am J Clin Pathol. 2007. PMID: 17638666
-
Comparison of hepatitis C virus subtyping by ns5b sequencing with 5'utr based methods.Minerva Med. 2012 Aug;103(4):293-7. Minerva Med. 2012. PMID: 22805621
-
Comparison of Sanger sequencing for hepatitis C virus genotyping with a commercial line probe assay in a tertiary hospital.BMC Infect Dis. 2019 Aug 22;19(1):738. doi: 10.1186/s12879-019-4386-4. BMC Infect Dis. 2019. PMID: 31438880 Free PMC article.
-
Genotyping of hepatitis C virus isolates by a new line probe assay using sequence information from both the 5'untranslated and the core regions.J Virol Methods. 2007 Aug;143(2):153-60. doi: 10.1016/j.jviromet.2007.03.006. Epub 2007 Apr 25. J Virol Methods. 2007. PMID: 17462747
Cited by
-
Serendipitous identification of natural intergenotypic recombinants of hepatitis C in Ireland.Virol J. 2006 Nov 15;3:95. doi: 10.1186/1743-422X-3-95. Virol J. 2006. PMID: 17107614 Free PMC article.
-
Improvement of hepatitis C virus (HCV) genotype determination with the new version of the INNO-LiPA HCV assay.J Clin Microbiol. 2007 Apr;45(4):1140-5. doi: 10.1128/JCM.01982-06. Epub 2007 Jan 24. J Clin Microbiol. 2007. PMID: 17251399 Free PMC article.
-
Performance comparison of the versant HCV genotype 2.0 assay (LiPA) and the abbott realtime HCV genotype II assay for detecting hepatitis C virus genotype 6.J Clin Microbiol. 2014 Oct;52(10):3685-92. doi: 10.1128/JCM.00882-14. Epub 2014 Aug 6. J Clin Microbiol. 2014. PMID: 25100817 Free PMC article.
-
Evaluation of Versant hepatitis C virus genotype assay (LiPA) 2.0.J Clin Microbiol. 2008 Jun;46(6):1901-6. doi: 10.1128/JCM.02390-07. Epub 2008 Apr 9. J Clin Microbiol. 2008. PMID: 18400913 Free PMC article.
-
Hepatitis C genotype 6: A concise review and response-guided therapy proposal.World J Hepatol. 2013 Sep 27;5(9):496-504. doi: 10.4254/wjh.v5.i9.496. World J Hepatol. 2013. PMID: 24073301 Free PMC article. Review.
References
-
- Buoro, S., S. Pizzighella, R. Boschetto, L. Pellizzari, M. Cusan, R. Bonaguro, C. Mengoli, C. Caudai, M. Padula, P. Egisto-Valensin, and G. Palu. 1999. Typing of hepatitis C virus by a new method based on restriction fragment length polymorphism. Intervirology 42:1-8. - PubMed
-
- Chinchai, T., J. Labout, S. Noppornpanth, A. Theamboonlers, B. L. Haagmans, A. D. Osterhaus, and Y. Poovorawan. 2003. Comparative study of different methods to genotype hepatitis C virus type 6 variants. J. Virol. Methods 109:195-201. - PubMed
-
- Dev, A. T., R. McCaw, V. Sundararajan, S. Bowden, and W. Sievert. 2002. Southeast Asian patients with chronic hepatitis C: the impact of novel genotypes and race on treatment outcome. Hepatology 36:1259-1265. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

