Assessment of genes associated with Streptococcus mutans biofilm morphology
- PMID: 16957255
- PMCID: PMC1563623
- DOI: 10.1128/AEM.00614-06
Assessment of genes associated with Streptococcus mutans biofilm morphology
Abstract
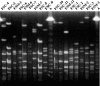
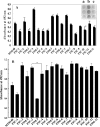
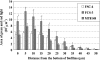
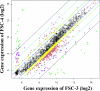
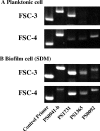
Streptococcus mutans, the major pathogen responsible for dental caries in humans, is a biofilm-forming bacterium. In the present study, 17 different pulsed-field gel electrophoresis patterns of genomic DNA were identified in S. mutans organisms isolated clinically from whole saliva. The S. mutans isolates showed different abilities to form biofilms on polystyrene surfaces in semidefined minimal medium cultures. Following cultivation in a flow cell system in tryptic soy broth with 0.25% sucrose and staining using a BacLight LIVE/DEAD system, two strains, designated FSC-3 and FSC-4, showed the greatest and least, respectively, levels of biofilm formation when examined with confocal laser scanning microscopy. Further, image analyses of spatial distribution and architecture were performed to quantify the merged green (live cells) and red (dead cells) light. The light intensity of the FSC-3 biofilm was greater than that of the FSC-4 biofilm in the bottom area but not in the top area. S. mutans whole-genome array results showed that approximately 3.8% of the genes were differentially expressed in the two strains, of which approximately 2.2%, including bacitracin transport ATP-binding protein gene glrA and a BLpL-like putative immunity protein gene, were activated in FSC-3. In addition, about 1.6% of the genes, including those associated with phosphotransferase system genes, were repressed. Analyses of the glrA-deficient strains and reverse transcription-PCR confirmed the role of the gene in biofilm formation. Differential assessment of biofilm-associated genes in clinical strains may provide useful information for understanding the morphological development of streptococcal biofilm, as well as for colonization of S. mutans.
Figures

References
-
- Ajdic, D., W. M. McShan, R. E. McLaughlin, G. Savic, J. Chang, M. B. Carson, C. Primeaux, R. Tian, S. Kenton, H. Jia, S. Lin, Y. Qian, S. Li, H. Zhu, F. Najar, H. Lai, J. White, B. A. Roe, and J. J. Ferretti. 2002. Genome sequence of Streptococcus mutans UA159, a cariogenic dental pathogen. Proc. Natl. Acad. Sci. USA 99:14434-14439. - PMC - PubMed
-
- Auschill, T. M., N. B. Arweiler, L. Netuschil, M. Brecx, E. Reich, and A. Sculean. 2001. Spatial distribution of vital and dead microorganisms in dental biofilms. Arch. Oral Biol. 46:471-476. - PubMed
-
- Berkowitz, R. J., and H. V. Jordan. 1975. Similarity of bacteriocins of Streptococcus mutans from mother and infant. Arch. Oral Biol. 20:725-730. - PubMed
-
- Bhagwat, S. P., J. Nary, and R. A. Burne. 2001. Effects of mutating putative two-component systems on biofilm formation by Streptococcus mutans UA159. FEMS Microbiol. Lett. 205:225-230. - PubMed
-
- Bolstad, B., R. Irizarry, M. Astrand, and T. Speed. 2003. A comparison of normalization methods for high density oligonucleotide array data based on bias and variance. Bioinfomatics 19:185-193. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

