The catalytic subunit of the proteasome is engaged in the entire process of estrogen receptor-regulated transcription
- PMID: 16957778
- PMCID: PMC1570434
- DOI: 10.1038/sj.emboj.7601306
The catalytic subunit of the proteasome is engaged in the entire process of estrogen receptor-regulated transcription
Abstract
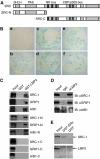
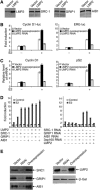
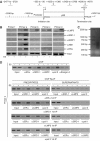
The ubiquitin-proteasome system plays an important role in a variety of cellular functions by means of its proteolytic activity. Interestingly, recent studies have indicated that the proteasome components are also integral parts of transcription complexes. In genome-wide screening for steroid receptor coactivator (SRC)-interacting proteins using yeast two-hybrid system, we found that the 20S proteasome beta subunit LMP2 (Low Molecular mass Polypeptide 2) interacts directly with the SRC coactivators. We showed that LMP2 is required for estrogen receptor (ER)-mediated gene transcription and for estrogen-stimulated cell cycle progression. We found that LMP2-associated proteasome is recruited to the entire sequence of ER target genes, implicating a role for the proteasome in both transcription initiation and elongation. We demonstrated that the recruitment of LMP2 by SRC coactivators is necessary for cyclic association of ER-regulated transcription complexes on ER targets. These results revealed a mechanism by which the proteasome machinery is recruited in ER-mediated gene transcription. Our experiments also provided evidence implicating SRC coactivators in gene transcription elongation.
Figures




References
-
- Anzick SL, Kononen J, Walker RL, Azorsa DO, Tanner MM, Guan XY, Sauter G, Kallioniemi OP, Trent JM, Meltzer PS (1997) AIB1, a steroid receptor coactivator amplified in breast and ovarian cancer. Science 277: 965–968 - PubMed
-
- Arndt K, Winston F (2005) An unexpected role for ubiquitylation of a transcriptional activator. Cell 120: 733–734 - PubMed
-
- Baker SP, Grant PA (2005) The proteasome: not just degrading anymore. Cell 123: 361–363 - PubMed
-
- Belandia B, Parker MG (2000) Functional interaction between the p160 coactivator proteins and the transcriptional enhancer factor family of transcription factors. J Biol Chem 275: 30801–30805 - PubMed
-
- Brummelkamp TR, Bernards R, Agami R (2002) A system for stable expression of short interfering RNAs in mammalian cells. Science 296: 550–553 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

