Genetic analysis of the GRIK2 modifier effect in Huntington's disease
- PMID: 16959037
- PMCID: PMC1618398
- DOI: 10.1186/1471-2202-7-62
Genetic analysis of the GRIK2 modifier effect in Huntington's disease
Abstract
Background: In Huntington's disease (HD), age at neurological onset is inversely correlated with the length of the CAG trinucleotide repeat mutation, but can be modified by genetic factors beyond the HD gene. Association of a relatively infrequent 16 TAA allele of a trinucleotide repeat polymorphism in the GRIK2 3'UTR with earlier than expected age at neurological onset has been suggested to reflect linkage disequilibrium with a functional polymorphism in GRIK2 or an adjacent gene.
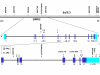
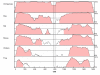
Results: We have tested this hypothesis by sequencing all GRIK2 exons, the exon-flanking sequences and 3'UTR in several individuals who were crucial to demonstrating the modifier effect, as they showed much earlier age at neurological onset than would be expected from the length of their HD CAG mutation. Though ten known SNPs were detected, no sequence variants were found in coding or adjacent sequence that could explain the modifier effect by linkage disequilibrium with the 16 TAA allele. Haplotype analysis using microsatellites, known SNPs and new variants discovered in the 3'UTR argues against a common ancestral origin for the 16 TAA repeat alleles in these individuals.
Conclusion: These data suggest that the modifier effect is actually due to the TAA repeat itself, possibly via a functional consequence on the GRIK2 mRNA.
Figures

References
-
- Gusella JF, Macdonald ME. Huntington's disease: seeing the pathogenic process through a genetic lens. Trends Biochem Sci. 2006 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

