Genetics of Streptomyces rimosus, the oxytetracycline producer
- PMID: 16959966
- PMCID: PMC1594589
- DOI: 10.1128/MMBR.00004-06
Genetics of Streptomyces rimosus, the oxytetracycline producer
Abstract
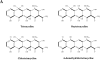
From a genetic standpoint, Streptomyces rimosus is arguably the best-characterized industrial streptomycete as the producer of oxytetracycline and other tetracycline antibiotics. Although resistance to these antibiotics has reduced their clinical use in recent years, tetracyclines have an increasing role in the treatment of emerging infections and noninfective diseases. Procedures for in vivo and in vitro genetic manipulations in S. rimosus have been developed since the 1950s and applied to study the genetic instability of S. rimosus strains and for the molecular cloning and characterization of genes involved in oxytetracycline biosynthesis. Recent advances in the methodology of genome sequencing bring the realistic prospect of obtaining the genome sequence of S. rimosus in the near term.
Figures



Similar articles
-
Molecular cloning of resistance genes and architecture of a linked gene cluster involved in biosynthesis of oxytetracycline by Streptomyces rimosus.Mol Gen Genet. 1989 Jan;215(2):231-8. doi: 10.1007/BF00339722. Mol Gen Genet. 1989. PMID: 2710100
-
Oxytetracycline hyper-production through targeted genome reduction of Streptomyces rimosus.mSystems. 2024 May 16;9(5):e0025024. doi: 10.1128/msystems.00250-24. Epub 2024 Apr 2. mSystems. 2024. PMID: 38564716 Free PMC article.
-
Reference-Grade Genome and Large Linear Plasmid of Streptomyces rimosus: Pushing the Limits of Nanopore Sequencing.Microbiol Spectr. 2022 Apr 27;10(2):e0243421. doi: 10.1128/spectrum.02434-21. Epub 2022 Apr 4. Microbiol Spectr. 2022. PMID: 35377231 Free PMC article.
-
Characterization of an oxytetracycline-resistance gene, otrA, of Streptomyces rimosus.Mol Microbiol. 1991 Dec;5(12):2923-33. doi: 10.1111/j.1365-2958.1991.tb01852.x. Mol Microbiol. 1991. PMID: 1809836
-
Genetics of antibiotic production in Streptomyces coelicolor A3(2), a model streptomycete.Biotechnology. 1995;28:65-102. doi: 10.1016/b978-0-7506-9095-9.50009-5. Biotechnology. 1995. PMID: 8688641 Review. No abstract available.
Cited by
-
Oxytetracycline biosynthesis.J Biol Chem. 2010 Sep 3;285(36):27509-15. doi: 10.1074/jbc.R110.130419. Epub 2010 Jun 3. J Biol Chem. 2010. PMID: 20522541 Free PMC article. Review.
-
Within-Species Genomic Variation and Variable Patterns of Recombination in the Tetracycline Producer Streptomyces rimosus.Front Microbiol. 2019 Mar 21;10:552. doi: 10.3389/fmicb.2019.00552. eCollection 2019. Front Microbiol. 2019. PMID: 30949149 Free PMC article.
-
Biosynthesis of Polyketides in Streptomyces.Microorganisms. 2019 May 6;7(5):124. doi: 10.3390/microorganisms7050124. Microorganisms. 2019. PMID: 31064143 Free PMC article. Review.
-
Biodiversity and Bioactive Potential of Actinomycetes from Unexplored High Altitude Regions of Kargil, India.Indian J Microbiol. 2024 Mar;64(1):110-124. doi: 10.1007/s12088-023-01133-1. Epub 2023 Dec 11. Indian J Microbiol. 2024. PMID: 38468743 Free PMC article.
-
Taxonomy, Physiology, and Natural Products of Actinobacteria.Microbiol Mol Biol Rev. 2015 Nov 25;80(1):1-43. doi: 10.1128/MMBR.00019-15. Print 2016 Mar. Microbiol Mol Biol Rev. 2015. PMID: 26609051 Free PMC article. Review.
References
-
- Ackermann, H.-W., and M. S. DuBow. 1987. Bacteriophage taxonomy, p. 13-28. In Viruses of prokaryotes, vol. 1. General properties of bacteriophages. CRC Press, Boca Raton, Fla.
-
- Ahel, I., D. Vujaklija, A. Mikoc, and V. Gamulin. 2002. Transcriptional analysis of the recA gene in Streptomyces rimosus: identification of the new type of promoter. FEMS Microbiol. Lett. 209:133-137. - PubMed
-
- Ahel, I., A. Mikoc, and V. Gamulin. 2005. recA gene expression in a streptomycete is mediated by the unusual C-terminus of RecA protein. FEMS Microbiol. Lett. 248:119-124. - PubMed
-
- Aigle, B., A. C. Holl, J. F. Angulo, P. Leblond, and B. Decaris. 1997. Characterization of two Streptomyces ambofaciens recA mutants: identification of the RecA protein by immunoblotting. FEMS Microbiol. Lett. 149:181-187. (Erratum, 155:131-132.) - PubMed
-
- Alačević, M. 1963. Interspecific recombination in Streptomyces. Nature 197:1323. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

