Mapping the pathways to staphylococcal pathogenesis by comparative secretomics
- PMID: 16959968
- PMCID: PMC1594592
- DOI: 10.1128/MMBR.00008-06
Mapping the pathways to staphylococcal pathogenesis by comparative secretomics
Abstract
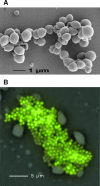
The gram-positive bacterium Staphylococcus aureus is a frequent component of the human microbial flora that can turn into a dangerous pathogen. As such, this organism is capable of infecting almost every tissue and organ system in the human body. It does so by actively exporting a variety of virulence factors to the cell surface and extracellular milieu. Upon reaching their respective destinations, these virulence factors have pivotal roles in the colonization and subversion of the human host. It is therefore of major importance to obtain a clear understanding of the protein transport pathways that are active in S. aureus. The present review aims to provide a state-of-the-art roadmap of staphylococcal secretomes, which include both protein transport pathways and the extracytoplasmic proteins of these organisms. Specifically, an overview is presented of the exported virulence factors, pathways for protein transport, signals for cellular protein retention or secretion, and the exoproteomes of different S. aureus isolates. The focus is on S. aureus, but comparisons with Staphylococcus epidermidis and other gram-positive bacteria, such as Bacillus subtilis, are included where appropriate. Importantly, the results of genomic and proteomic studies on S. aureus secretomes are integrated through a comparative "secretomics" approach, resulting in the first definition of the core and variant secretomes of this bacterium. While the core secretome seems to be largely employed for general housekeeping functions which are necessary to thrive in particular niches provided by the human host, the variant secretome seems to contain the "gadgets" that S. aureus needs to conquer these well-protected niches.
Figures

References
-
- Adhikari, R. P., and R. P. Novick. 2005. Subinhibitory cerulenin inhibits staphylococcal exoprotein production by blocking transcription rather than by blocking secretion. Microbiology 151:3059-3069. - PubMed
-
- Alami, M., I. Lüke, S. Deitermann, G. Eisner, H. G. Koch, J. Brunner, and M. Müller. 2003. Differential interactions between a twin-arginine signal peptide and its translocase in Escherichia coli. Mol. Cell 12:937-946. - PubMed
-
- Antelmann, H., H. Tjalsma, B. Voigt, S. Ohlmeier, S. Bron, J. M. van Dijl, and M. Hecker. 2001. A proteomic view on genome-based signal peptide predictions. Genome Res. 11:1484-1502. - PubMed
-
- Avison, M. B., P. M. Bennett, R. A. Howe, and T. R. Walsh. 2002. Preliminary analysis of the genetic basis for vancomycin resistance in Staphylococcus aureus strain Mu50. J. Antimicrob. Chemother. 49:255-260. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

