Nuclear ADP-ribosylation reactions in mammalian cells: where are we today and where are we going?
- PMID: 16959969
- PMCID: PMC1594587
- DOI: 10.1128/MMBR.00040-05
Nuclear ADP-ribosylation reactions in mammalian cells: where are we today and where are we going?
Abstract
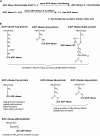
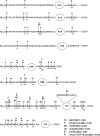
Since poly-ADP ribose was discovered over 40 years ago, there has been significant progress in research into the biology of mono- and poly-ADP-ribosylation reactions. During the last decade, it became clear that ADP-ribosylation reactions play important roles in a wide range of physiological and pathophysiological processes, including inter- and intracellular signaling, transcriptional regulation, DNA repair pathways and maintenance of genomic stability, telomere dynamics, cell differentiation and proliferation, and necrosis and apoptosis. ADP-ribosylation reactions are phylogenetically ancient and can be classified into four major groups: mono-ADP-ribosylation, poly-ADP-ribosylation, ADP-ribose cyclization, and formation of O-acetyl-ADP-ribose. In the human genome, more than 30 different genes coding for enzymes associated with distinct ADP-ribosylation activities have been identified. This review highlights the recent advances in the rapidly growing field of nuclear mono-ADP-ribosylation and poly-ADP-ribosylation reactions and the distinct ADP-ribosylating enzyme families involved in these processes, including the proposed family of novel poly-ADP-ribose polymerase-like mono-ADP-ribose transferases and the potential mono-ADP-ribosylation activities of the sirtuin family of NAD(+)-dependent histone deacetylases. A special focus is placed on the known roles of distinct mono- and poly-ADP-ribosylation reactions in physiological processes, such as mitosis, cellular differentiation and proliferation, telomere dynamics, and aging, as well as "programmed necrosis" (i.e., high-mobility-group protein B1 release) and apoptosis (i.e., apoptosis-inducing factor shuttling). The proposed molecular mechanisms involved in these processes, such as signaling, chromatin modification (i.e., "histone code"), and remodeling of chromatin structure (i.e., DNA damage response, transcriptional regulation, and insulator function), are described. A potential cross talk between nuclear ADP-ribosylation processes and other NAD(+)-dependent pathways is discussed.
Figures




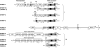

References
-
- Adamietz, P., and A. Rudolph. 1984. ADP-ribosylation of nuclear proteins in vivo. Identification of histone H2B as a major acceptor for mono- and poly(ADP-ribose) in dimethyl sulfate-treated hepatoma AH 7974 cells. J. Biol. Chem. 259:6841-6846. - PubMed
-
- Adebanjo, O. A., H. K. Anandatheerthavarada, A. P. Koval, B. S. Moonga, G. Biswas, L. Sun, B. R. Sodam, P. J. Bevis, C. L. Huang, S. Epstein, F. A. Lai, N. G. Avadhani, and M. Zaidi. 1999. A new function for CD38/ADP-ribosyl cyclase in nuclear Ca2+ homeostasis. Nat. Cell Biol. 1:409-414. - PubMed
-
- Adolph, K. W., and M. K. Song. 1985. Variations in ADP-ribosylation of nuclear scaffold proteins during the HeLa cell cycle. Biochem. Biophys. Res. Commun. 126:840-847. - PubMed
-
- Adriouch, S., W. Ohlrogge, F. Haag, F. Koch-Nolte, and M. Seman. 2001. Rapid induction of naive T cell apoptosis by ecto-nicotinamide adenine dinucleotide: requirement for mono(ADP-ribosyl)transferase 2 and a downstream effector. J. Immunol. 167:196-203. - PubMed
-
- Aguiar, R. C., K. Takeyama, C. He, K. Kreinbrink, and M. A. Shipp. 2005. B-aggressive lymphoma family proteins have unique domains that modulate transcription and exhibit poly(ADP-ribose) polymerase activity. J. Biol. Chem. 280:33756-33765. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

