Epigenetic gene regulation in the bacterial world
- PMID: 16959970
- PMCID: PMC1594586
- DOI: 10.1128/MMBR.00016-06
Epigenetic gene regulation in the bacterial world
Abstract
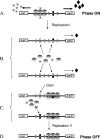
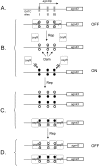
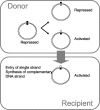
Like many eukaryotes, bacteria make widespread use of postreplicative DNA methylation for the epigenetic control of DNA-protein interactions. Unlike eukaryotes, however, bacteria use DNA adenine methylation (rather than DNA cytosine methylation) as an epigenetic signal. DNA adenine methylation plays roles in the virulence of diverse pathogens of humans and livestock animals, including pathogenic Escherichia coli, Salmonella, Vibrio, Yersinia, Haemophilus, and Brucella. In Alphaproteobacteria, methylation of adenine at GANTC sites by the CcrM methylase regulates the cell cycle and couples gene transcription to DNA replication. In Gammaproteobacteria, adenine methylation at GATC sites by the Dam methylase provides signals for DNA replication, chromosome segregation, mismatch repair, packaging of bacteriophage genomes, transposase activity, and regulation of gene expression. Transcriptional repression by Dam methylation appears to be more common than transcriptional activation. Certain promoters are active only during the hemimethylation interval that follows DNA replication; repression is restored when the newly synthesized DNA strand is methylated. In the E. coli genome, however, methylation of specific GATC sites can be blocked by cognate DNA binding proteins. Blockage of GATC methylation beyond cell division permits transmission of DNA methylation patterns to daughter cells and can give rise to distinct epigenetic states, each propagated by a positive feedback loop. Switching between alternative DNA methylation patterns can split clonal bacterial populations into epigenetic lineages in a manner reminiscent of eukaryotic cell differentiation. Inheritance of self-propagating DNA methylation patterns governs phase variation in the E. coli pap operon, the agn43 gene, and other loci encoding virulence-related cell surface functions.
Figures




References
-
- Aoki, S. K., R. Pamma, A. D. Hernday, J. E. Bickham, B. A. Braaten, and D. A. Low. 2005. Contact-dependent inhibition of growth in Escherichia coli. Science 309:1245-1248. - PubMed
-
- Appelmelk, B. J., S. L. Martin, M. A. Monteiro, C. A. Clayton, A. A. McColm, P. Zheng, T. Verboom, J. J. Maaskant, D. H. van den Eijnden, C. H. Hokke, M. B. Perry, C. M. Vandenbroucke-Grauls, and J. G. Kusters. 1999. Phase variation in Helicobacter pylori lipopolysaccharide due to changes in the lengths of poly(C) tracts in α3-fucosyltransferase genes. Infect. Immun. 67:5361-5366. - PMC - PubMed
-
- Arber, W. 1974. DNA modification and restriction. Prog. Nucleic Acid Res. Mol. Biol. 14:1-37. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

