Global gene expression during stringent response in Corynebacterium glutamicum in presence and absence of the rel gene encoding (p)ppGpp synthase
- PMID: 16961923
- PMCID: PMC1578569
- DOI: 10.1186/1471-2164-7-230
Global gene expression during stringent response in Corynebacterium glutamicum in presence and absence of the rel gene encoding (p)ppGpp synthase
Abstract
Background: The stringent response is the initial reaction of microorganisms to nutritional stress. During stringent response the small nucleotides (p)ppGpp act as global regulators and reprogram bacterial transcription. In this work, the genetic network controlled by the stringent response was characterized in the amino acid-producing Corynebacterium glutamicum.
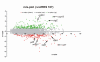
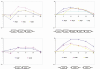
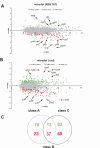
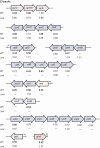
Results: The transcriptome of a C. glutamicum rel gene deletion mutant, unable to synthesize (p)ppGpp and to induce the stringent response, was compared with that of its rel-proficient parent strain by microarray analysis. A total of 357 genes were found to be transcribed differentially in the rel-deficient mutant strain. In a second experiment, the stringent response was induced by addition of DL-serine hydroxamate (SHX) in early exponential growth phase. The time point of the maximal effect on transcription was determined by real-time RT-PCR using the histidine and serine biosynthetic genes. Transcription of all of these genes reached a maximum at 10 minutes after SHX addition. Microarray experiments were performed comparing the transcriptomes of SHX-induced cultures of the rel-proficient strain and the rel mutant. The differentially expressed genes were grouped into three classes. Class A comprises genes which are differentially regulated only in the presence of an intact rel gene. This class includes the non-essential sigma factor gene sigB which was upregulated and a large number of genes involved in nitrogen metabolism which were downregulated. Class B comprises genes which were differentially regulated in response to SHX in both strains, independent of the rel gene. A large number of genes encoding ribosomal proteins fall into this class, all being downregulated. Class C comprises genes which were differentially regulated in response to SHX only in the rel mutant. This class includes genes encoding putative stress proteins and global transcriptional regulators that might be responsible for the complex transcriptional patterns detected in the rel mutant when compared directly with its rel-proficient parent strain.
Conclusion: In C. glutamicum the stringent response enfolds a fast answer to an induced amino acid starvation on the transcriptome level. It also showed some significant differences to the transcriptional reactions occurring in Escherichia coli and Bacillus subtilis. Notable are the rel-dependent regulation of the nitrogen metabolism genes and the rel-independent regulation of the genes encoding ribosomal proteins.
Figures

Similar articles
-
Physiological analysis of the stringent response elicited in an extreme thermophilic bacterium, Thermus thermophilus.J Bacteriol. 2006 Oct;188(20):7111-22. doi: 10.1128/JB.00574-06. J Bacteriol. 2006. PMID: 17015650 Free PMC article.
-
The alternative sigma factor SigB of Corynebacterium glutamicum modulates global gene expression during transition from exponential growth to stationary phase.BMC Genomics. 2007 Jan 4;8:4. doi: 10.1186/1471-2164-8-4. BMC Genomics. 2007. PMID: 17204139 Free PMC article.
-
The extracytoplasmic function-type sigma factor SigM of Corynebacterium glutamicum ATCC 13032 is involved in transcription of disulfide stress-related genes.J Bacteriol. 2007 Jul;189(13):4696-707. doi: 10.1128/JB.00382-07. Epub 2007 May 4. J Bacteriol. 2007. PMID: 17483229 Free PMC article.
-
Direct binding targets of the stringent response alarmone (p)ppGpp.Mol Microbiol. 2012 Sep;85(6):1029-43. doi: 10.1111/j.1365-2958.2012.08177.x. Epub 2012 Aug 2. Mol Microbiol. 2012. PMID: 22812515 Review.
-
[The stringent response--bacterial mechanism of an adaptive stress response].Postepy Biochem. 2006;52(1):87-93. Postepy Biochem. 2006. PMID: 16869306 Review. Polish.
Cited by
-
Histidine biosynthesis, its regulation and biotechnological application in Corynebacterium glutamicum.Microb Biotechnol. 2014 Jan;7(1):5-25. doi: 10.1111/1751-7915.12055. Epub 2013 Apr 25. Microb Biotechnol. 2014. PMID: 23617600 Free PMC article. Review.
-
Global Gene Expression Analysis of Cross-Protected Phenotype of Pectobacterium atrosepticum.PLoS One. 2017 Jan 12;12(1):e0169536. doi: 10.1371/journal.pone.0169536. eCollection 2017. PLoS One. 2017. PMID: 28081189 Free PMC article.
-
Transcriptomic analysis of three Veillonella spp. present in carious dentine and in the saliva of caries-free individuals.Front Cell Infect Microbiol. 2015 Mar 26;5:25. doi: 10.3389/fcimb.2015.00025. eCollection 2015. Front Cell Infect Microbiol. 2015. PMID: 25859434 Free PMC article.
-
Factors enhancing L-valine production by the growth-limited L-isoleucine auxotrophic strain Corynebacterium glutamicum DeltailvA DeltapanB ilvNM13 (pECKAilvBNC).J Ind Microbiol Biotechnol. 2010 Jul;37(7):689-99. doi: 10.1007/s10295-010-0712-y. Epub 2010 Apr 4. J Ind Microbiol Biotechnol. 2010. PMID: 20364396
-
Stress response regulators identified through genome-wide transcriptome analysis of the (p)ppGpp-dependent response in Rhizobium etli.Genome Biol. 2011;12(2):R17. doi: 10.1186/gb-2011-12-2-r17. Epub 2011 Feb 16. Genome Biol. 2011. PMID: 21324192 Free PMC article.
References
-
- Cashel M, Gentry DR, Hernandez VJ, Villa D. In: The stringent response. Neidhardt FC, Curtiss R III, Ingraham JL, Lin ECCK, editor. 1996. pp. 1458–1496.
-
- Strauch E, Takano E, Baylis HA, Bibb MJ. The stringent response in Streptomyces coelicolor A3(2) Mol Microbiol. 1991;5:289–298. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

