Sites of interaction of a precursor polypeptide on the export chaperone SecB mapped by site-directed spin labeling
- PMID: 16962134
- PMCID: PMC2925277
- DOI: 10.1016/j.jmb.2006.07.021
Sites of interaction of a precursor polypeptide on the export chaperone SecB mapped by site-directed spin labeling
Abstract
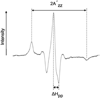
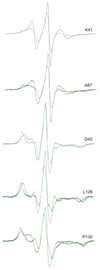
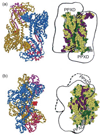
Export of protein into the periplasm of Escherichia coli via the general secretory system requires that the transported polypeptides be devoid of stably folded tertiary structure. Capture of the precursor polypeptides before they fold is achieved by the promiscuous binding to the chaperone SecB. SecB delivers its ligand to export sites through its specific binding to SecA, a peripheral component of the membrane translocon. At the translocon the ligand is passed from SecB to SecA and subsequently through the SecYEG channel. We have previously used site-directed spin labeling and electron paramagnetic resonance spectroscopy to establish a docking model between SecB and SecA. Here we report use of the same strategy to map the pathway of a physiologic ligand, the unfolded form of precursor galactose-binding protein, on SecB. Our set of SecB variants each containing a single cysteine, which was used in the previous study, has been expanded to 48 residues, which cover 49% of the surface of SecB. The residues on SecB involved in contacts were identified as those that, upon addition of the unfolded polypeptide ligand, showed changes in spectral line shape consistent with restricted motion of the nitroxide. We conclude that the bound precursor makes contact with a large portion of the surface of the small chaperone. The sites on SecB that interact with the ligand are compared with the previously identified sites that interact with SecA and a model for transfer of the ligand is discussed.
Figures








References
-
- Berks BC, Sargent F, Palmer T. The Tat protein export pathway. Mol. Microbiol. 2000;35:260–274. - PubMed
-
- Economou A. Bacterial secretome: the assembly manual and operating instructions. Molecular Membrane Biology. 2002;19:159–169. - PubMed
-
- Randall LL, Hardy SJS. High selectivity with low specificity: how SecB has solved the paradox of chaperone binding. Trends Biochem. Sci. 1995;20:65–69. - PubMed
-
- Crane JM, Mao C, Lilly AA, Smith VF, Suo Y, Hubbell WL, Randall LL. Mapping of the docking of SecA onto the chaperone SecB by site-directed spin labeling: insight into the mechanism of ligand transfer during protein export. J. Mol. Biol. 2005;353:295–307. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

