A specific loop in human DNA polymerase mu allows switching between creative and DNA-instructed synthesis
- PMID: 16963491
- PMCID: PMC1636348
- DOI: 10.1093/nar/gkl457
A specific loop in human DNA polymerase mu allows switching between creative and DNA-instructed synthesis
Abstract
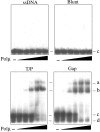
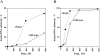
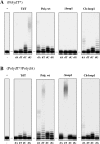
Human DNA polymerase mu (Polmu) is a family X member that has terminal transferase activity but, in spite of a non-orthodox selection of the template information, displays its maximal catalytic efficiency in DNA-templated reactions. As terminal deoxynucleotidyl transferase (TdT), Polmu has a specific loop (loop1) that could provide this enzyme with its terminal transferase activity. When loop1 was deleted, human Polmu lacked TdT activity but improved DNA-binding and DNA template-dependent polymerization. Interestingly, when loop1 from TdT was inserted in Polmu (substituting its cognate loop1), the resulting chimaera displayed TdT activity, preferentially inserting dGTP residues, but had a strongly reduced template-dependent polymerization activity. Therefore, a specialized loop in Polmu, that could adopt alternative conformations, appears to provide this enzyme with a dual capacity: (i) template independency to create new DNA information, in which loop1 would have an active role by acting as a 'pseudotemplate'; (ii) template-dependent polymerization, in which loop1 must allow binding of the template strand. Recent in vivo and in vitro data suggest that such a dual capacity could be advantageous to resolve microhomology-mediated end-joining reactions.
Figures


Similar articles
-
Conferring a template-dependent polymerase activity to terminal deoxynucleotidyltransferase by mutations in the Loop1 region.Nucleic Acids Res. 2009 Aug;37(14):4642-56. doi: 10.1093/nar/gkp460. Epub 2009 Jun 5. Nucleic Acids Res. 2009. PMID: 19502493 Free PMC article.
-
Structural evidence for an in trans base selection mechanism involving Loop1 in polymerase μ at an NHEJ double-strand break junction.J Biol Chem. 2019 Jul 5;294(27):10579-10595. doi: 10.1074/jbc.RA119.008739. Epub 2019 May 28. J Biol Chem. 2019. PMID: 31138645 Free PMC article.
-
Limited terminal transferase in human DNA polymerase mu defines the required balance between accuracy and efficiency in NHEJ.Proc Natl Acad Sci U S A. 2009 Sep 22;106(38):16203-8. doi: 10.1073/pnas.0908492106. Epub 2009 Sep 4. Proc Natl Acad Sci U S A. 2009. PMID: 19805281 Free PMC article.
-
Terminal Deoxynucleotidyl Transferase in the Synthesis and Modification of Nucleic Acids.Chembiochem. 2019 Apr 1;20(7):860-871. doi: 10.1002/cbic.201800658. Epub 2019 Jan 25. Chembiochem. 2019. PMID: 30451377 Review.
-
DNA polymerase mu, a candidate hypermutase?Philos Trans R Soc Lond B Biol Sci. 2001 Jan 29;356(1405):99-109. doi: 10.1098/rstb.2000.0754. Philos Trans R Soc Lond B Biol Sci. 2001. PMID: 11205337 Free PMC article. Review.
Cited by
-
Essential role for polymerase specialization in cellular nonhomologous end joining.Proc Natl Acad Sci U S A. 2015 Aug 18;112(33):E4537-45. doi: 10.1073/pnas.1505805112. Epub 2015 Aug 3. Proc Natl Acad Sci U S A. 2015. PMID: 26240371 Free PMC article.
-
DNA expansions generated by human Polμ on iterative sequences.Nucleic Acids Res. 2013 Jan 7;41(1):253-63. doi: 10.1093/nar/gks1054. Epub 2012 Nov 9. Nucleic Acids Res. 2013. PMID: 23143108 Free PMC article.
-
Characterization of terminal deoxynucleotidyl transferase and polymerase mu in zebrafish.Immunogenetics. 2007 Sep;59(9):735-44. doi: 10.1007/s00251-007-0241-7. Epub 2007 Aug 14. Immunogenetics. 2007. PMID: 17701034
-
The X family portrait: structural insights into biological functions of X family polymerases.DNA Repair (Amst). 2007 Dec 1;6(12):1709-25. doi: 10.1016/j.dnarep.2007.05.009. Epub 2007 Jul 12. DNA Repair (Amst). 2007. PMID: 17631059 Free PMC article. Review.
-
Molecular Dynamics Simulations and Structural Analysis to Decipher Functional Impact of a Twenty Residue Insert in the Ternary Complex of Mus musculus TdT Isoform.PLoS One. 2016 Jun 16;11(6):e0157286. doi: 10.1371/journal.pone.0157286. eCollection 2016. PLoS One. 2016. PMID: 27311013 Free PMC article.
References
-
- Beard W.A., Wilson S.H. Structural design of a eukaryotic DNA repair polymerase: DNA polymerase beta. Mutat. Res. 2000;460:231–244. - PubMed
-
- Garcia-Diaz M., Bebenek K., Krahn J.M., Blanco L., Kunkel T.A., Pedersen L.C. A structural solution for the DNA polymerase lambda-dependent repair of DNA gaps with minimal homology. Mol. Cell. 2004;13:561–572. - PubMed
-
- Garcia-Diaz M., Bebenek K., Krahn J.M., Kunkel T.A., Pedersen L.C. A closed conformation for the Pol lambda catalytic cycle. Nature Struct. Mol. Biol. 2005;12:97–98. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

