Characterization of DGCR8/Pasha, the essential cofactor for Drosha in primary miRNA processing
- PMID: 16963499
- PMCID: PMC1636349
- DOI: 10.1093/nar/gkl458
Characterization of DGCR8/Pasha, the essential cofactor for Drosha in primary miRNA processing
Abstract
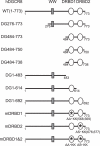
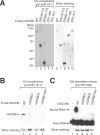
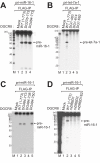
DGCR8/Pasha is an essential cofactor for Drosha, a nuclear RNase III that cleaves the local hairpin structures embedded in long primary microRNA transcripts (pri-miRNAs) in eukaryotes. Although our knowledge of pri-miRNA processing has significantly advanced in recent years, the precise role of DGCR8 in this pathway remains unclear. In our present study, we dissect the domains in DGCR8 that contribute to the processing of pri-miRNAs and the subcellular localization of DGCR8. Drosha is stabilized through an interaction between its middle domain and the conserved C-terminal domain of DGCR8. Furthermore, DGCR8, but not Drosha, can directly and stably interact with pri-miRNAs, and the tandem dsRNA-binding domains (dsRBDs) in DGCR8 are responsible for this recognition. Moreover, the DGCR8 N-terminal region upstream of its dsRBDs is unnecessary for pri-miRNA processing but is critical for nuclear localization. Our study thus provides further insights into the mechanism of action of the Drosha-DGCR8 complex in pri-miRNA processing.
Figures



References
-
- Bartel D.P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. - PubMed
-
- Kim V.N. Small RNAs: Classification, Biogenesis, and Function. Mol. Cells. 2005;19:1–15. - PubMed
-
- Lewis B.P., Burge C.B., Bartel D.P. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. - PubMed
-
- Croce C.M., Calin G.A. miRNAs, cancer, and stem cell division. Cell. 2005;122:6–7. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

