Biochemical and molecular characterization of AtPAP26, a vacuolar purple acid phosphatase up-regulated in phosphate-deprived Arabidopsis suspension cells and seedlings
- PMID: 16963519
- PMCID: PMC1630754
- DOI: 10.1104/pp.106.087171
Biochemical and molecular characterization of AtPAP26, a vacuolar purple acid phosphatase up-regulated in phosphate-deprived Arabidopsis suspension cells and seedlings
Abstract
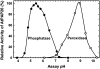
A vacuolar acid phosphatase (APase) that accumulates during phosphate (Pi) starvation of Arabidopsis (Arabidopsis thaliana) suspension cells was purified to homogeneity. The final preparation is a purple APase (PAP), as it exhibited a pink color in solution (A(max) = 520 nm). It exists as a 100-kD homodimer composed of 55-kD glycosylated subunits that cross-reacted with an anti-(tomato intracellular PAP)-IgG. BLAST analysis of its 23-amino acid N-terminal sequence revealed that this PAP is encoded by At5g34850 (AtPAP26; one of 29 PAP genes in Arabidopsis) and that a 30-amino acid signal peptide is cleaved from the AtPAP26 preprotein during its translocation into the vacuole. AtPAP26 displays much stronger sequence similarity to orthologs from other plants than to other Arabidopsis PAPs. AtPAP26 exhibited optimal activity at pH 5.6 and broad substrate selectivity. The 5-fold increase in APase activity that occurred in Pi-deprived cells was paralleled by a similar increase in the amount of a 55-kD anti-(tomato PAP or AtPAP26)-IgG immunoreactive polypeptide and a >30-fold reduction in intracellular free Pi concentration. Semiquantitative reverse transcription-PCR indicated that Pi-sufficient, Pi-starved, and Pi-resupplied cells contain similar amounts of AtPAP26 transcripts. Thus, transcriptional controls appear to exert little influence on AtPAP26 levels, relative to translational and/or proteolytic controls. APase activity and AtPAP26 protein levels were also up-regulated in shoots and roots of Pi-deprived Arabidopsis seedlings. We hypothesize that AtPAP26 recycles Pi from intracellular P metabolites in Pi-starved Arabidopsis. As AtPAP26 also exhibited alkaline peroxidase activity, a potential additional role in the metabolism of reactive oxygen species is discussed.
Figures




Similar articles
-
The dual-targeted purple acid phosphatase isozyme AtPAP26 is essential for efficient acclimation of Arabidopsis to nutritional phosphate deprivation.Plant Physiol. 2010 Jul;153(3):1112-22. doi: 10.1104/pp.110.153270. Epub 2010 Mar 26. Plant Physiol. 2010. PMID: 20348213 Free PMC article.
-
Biochemical and molecular characterization of AtPAP12 and AtPAP26: the predominant purple acid phosphatase isozymes secreted by phosphate-starved Arabidopsis thaliana.Plant Cell Environ. 2010 Nov;33(11):1789-803. doi: 10.1111/j.1365-3040.2010.02184.x. Plant Cell Environ. 2010. PMID: 20545876
-
The secreted purple acid phosphatase isozymes AtPAP12 and AtPAP26 play a pivotal role in extracellular phosphate-scavenging by Arabidopsis thaliana.J Exp Bot. 2012 Nov;63(18):6531-42. doi: 10.1093/jxb/ers309. Epub 2012 Nov 3. J Exp Bot. 2012. PMID: 23125358 Free PMC article.
-
Plant purple acid phosphatases - genes, structures and biological function.Acta Biochim Pol. 2003;50(4):1245-56. Acta Biochim Pol. 2003. PMID: 14740011 Review.
-
Metabolic adaptations of phosphate-starved plants.Plant Physiol. 2011 Jul;156(3):1006-15. doi: 10.1104/pp.111.175281. Epub 2011 May 11. Plant Physiol. 2011. PMID: 21562330 Free PMC article. Review. No abstract available.
Cited by
-
Purification and characterization of acid phosphatase from Macrotyloma uiflorum seeds.J Food Sci Technol. 2018 Jan;55(1):313-320. doi: 10.1007/s13197-017-2941-9. Epub 2017 Oct 23. J Food Sci Technol. 2018. PMID: 29358824 Free PMC article.
-
Allelic Variation in GmPAP14 Alters Gene Expression to Affect Acid Phosphatase Activity in Soybean.Int J Mol Sci. 2023 Mar 11;24(6):5398. doi: 10.3390/ijms24065398. Int J Mol Sci. 2023. PMID: 36982472 Free PMC article.
-
Molecular Mechanisms of Phosphorus Metabolism and Transport during Leaf Senescence.Plants (Basel). 2015 Dec 16;4(4):773-98. doi: 10.3390/plants4040773. Plants (Basel). 2015. PMID: 27135351 Free PMC article. Review.
-
Purple acid phosphatase 10c encodes a major acid phosphatase that regulates plant growth under phosphate-deficient conditions in rice.J Exp Bot. 2020 Jul 6;71(14):4321-4332. doi: 10.1093/jxb/eraa179. J Exp Bot. 2020. PMID: 32270183 Free PMC article.
-
The maize (Zea mays ssp. mays var. B73) genome encodes 33 members of the purple acid phosphatase family.Front Plant Sci. 2015 May 19;6:341. doi: 10.3389/fpls.2015.00341. eCollection 2015. Front Plant Sci. 2015. PMID: 26042133 Free PMC article.
References
-
- Amtmann A, Hammond JP, Armengaud P, White J (2006) Nutrient sensing in plants: potassium and phosphorus. Adv Bot Res 43: 209–257
-
- Bozzo GG, Dunn EL, Plaxton WC (2006) Differential synthesis of phosphate-starvation inducible purple acid phosphatase isozymes in tomato (Lycopersicon esculentum) suspension cells and seedlings. Plant Cell Environ 29: 303–313 - PubMed
-
- Bozzo GG, Raghothama KG, Plaxton WC (2002) Purification and characterization of two secreted purple acid phosphatase isozymes from phosphate-starved tomato (Lycopersicon esculentum) cell cultures. Eur J Biochem 269: 6278–6286 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

