Isolation of mtpim proves Tnt1 a useful reverse genetics tool in Medicago truncatula and uncovers new aspects of AP1-like functions in legumes
- PMID: 16963524
- PMCID: PMC1630737
- DOI: 10.1104/pp.106.083543
Isolation of mtpim proves Tnt1 a useful reverse genetics tool in Medicago truncatula and uncovers new aspects of AP1-like functions in legumes
Abstract
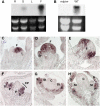
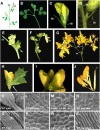
Comparative studies help shed light on how the huge diversity in plant forms found in nature has been produced. We use legume species to study developmental differences in inflorescence architecture and flower ontogeny with classical models such as Arabidopsis thaliana or Antirrhinum majus. Whereas genetic control of these processes has been analyzed mostly in pea (Pisum sativum), Medicago truncatula is emerging as a promising alternative system for these studies due to the availability of a range of genetic tools. To assess the use of the retrotransposon Tnt1 for reverse genetics in M. truncatula, we screened a small Tnt1-mutagenized population using degenerate primers for MADS-box genes, known controllers of plant development. We describe here the characterization of mtpim, a new mutant caused by the insertion of Tnt1 in a homolog to the PROLIFERATING INFLORESCENCE MERISTEM (PIM)/APETALA1 (AP1)/SQUAMOSA genes. mtpim shows flower-to-inflorescence conversion and altered flowers with sepals transformed into leaves, indicating that MtPIM controls floral meristem identity and flower development. Although more extreme, this phenotype resembles the pea pim mutants, supporting the idea that M. truncatula could be used to complement analysis of reproductive development already initiated in pea. In fact, our study reveals aspects not shown by analysis of pea mutants: that the mutation in the AP1 homolog interferes with the specification of floral organs from common primordia and causes conversion of sepals into leaves, in addition to true conversion of flowers into inflorescences. The isolation of mtpim represents a proof of concept demonstrating that Tnt1 populations can be efficiently used in reverse genetics screenings in M. truncatula.
Figures




References
-
- Azpiroz-Leehan R, Feldmann KA (1997) T-DNA insertion mutagenesis in Arabidopsis: going back and forth. Trends Genet 13: 152–156 - PubMed
-
- Benlloch R, Navarro C, Beltrán JP, Cañas LA (2002) Floral development of the model legume Medicago truncatula: ontogeny studies as a tool to better characterize homeotic mutations. Sex Plant Reprod 15: 231–241
-
- Berbel A, Navarro C, Ferrándiz C, Cañas LA, Madueño F, Beltrán JP (2001) Analysis of PEAM4, the pea AP1 functional homologue, supports a model for AP1-like genes controlling both floral meristem and floral organ identity in different plant species. Plant J 25: 441–451 - PubMed
-
- Bowman JL, Alvarez J, Weigel D, Meyerowitz EM, Smyth DR (1993) Control of flower development in Arabidopsis thaliana by APETALA1 and interacting genes. Development 119: 721–743
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

