Consistency of polyamine profiles and expression of arginine decarboxylase in mitosis during zygotic embryogenesis of Scots pine
- PMID: 16963525
- PMCID: PMC1630739
- DOI: 10.1104/pp.106.083030
Consistency of polyamine profiles and expression of arginine decarboxylase in mitosis during zygotic embryogenesis of Scots pine
Abstract
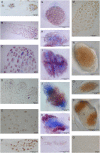
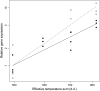
In this study, we show that both arginine decarboxylase (ADC) protein and mRNA transcript are present at different phases of mitosis in Scots pine (Pinus sylvestris) zygotic embryogenesis. We also examined the consistency of polyamine (PA) profiles with the effective temperature sum, the latter indicating the developmental stage of the embryos. PA metabolism was analyzed by fitting statistical regression models to the data of free and soluble conjugated PAs, to the enzyme activities of ADC and ornithine decarboxylase (ODC), as well as to the gene expression of ADC. According to the fitted models, PAs typically had the tendency to increase at the early stages but decrease at the late stages of embryogenesis. Only the free putrescine fraction remained stable during embryo development. The PA biosynthesis strongly preferred the ADC pathway. Both ADC gene expression and ADC enzyme activity were substantially higher than putative ODC gene expression or ODC enzyme activity, respectively. ADC gene expression and enzyme activity increased during embryogenesis, which suggests the involvement of transcriptional regulation in the expression of ADC. Both ADC mRNA and ADC protein localized in dividing cells of embryo meristems and more specifically within the mitotic spindle apparatus and close to the chromosomes, respectively. The results suggest the essential role of ADC in the mitosis of plant cells.
Figures


Similar articles
-
Moderate stress responses and specific changes in polyamine metabolism characterize Scots pine somatic embryogenesis.Tree Physiol. 2016 Mar;36(3):392-402. doi: 10.1093/treephys/tpv136. Epub 2016 Jan 19. Tree Physiol. 2016. PMID: 26786537 Free PMC article.
-
Changes in polyamine content and localization of Pinus sylvestris ADC and Suillus variegatus ODC mRNA transcripts during the formation of mycorrhizal interaction in an in vitro cultivation system.J Exp Bot. 2006;57(11):2795-804. doi: 10.1093/jxb/erl049. Epub 2006 Jul 25. J Exp Bot. 2006. PMID: 16868043
-
Elucidation of the polyamine biosynthesis pathway during Brazilian pine (Araucaria angustifolia) seed development.Tree Physiol. 2017 Jan 31;37(1):116-130. doi: 10.1093/treephys/tpw107. Tree Physiol. 2017. PMID: 28175909
-
The antizyme family for regulating polyamines.J Biol Chem. 2018 Nov 30;293(48):18730-18735. doi: 10.1074/jbc.TM118.003339. Epub 2018 Oct 24. J Biol Chem. 2018. PMID: 30355739 Free PMC article. Review.
-
Translational regulation of ornithine decarboxylase and other enzymes of the polyamine pathway.Int J Biochem Cell Biol. 1999 Jan;31(1):107-22. doi: 10.1016/s1357-2725(98)00135-6. Int J Biochem Cell Biol. 1999. PMID: 10216947 Review.
Cited by
-
Scots pine aminopropyltransferases shed new light on evolution of the polyamine biosynthesis pathway in seed plants.Ann Bot. 2018 May 11;121(6):1243-1256. doi: 10.1093/aob/mcy012. Ann Bot. 2018. PMID: 29462244 Free PMC article.
-
One tissue, two fates: different roles of megagametophyte cells during Scots pine embryogenesis.J Exp Bot. 2009;60(4):1375-86. doi: 10.1093/jxb/erp020. Epub 2009 Feb 26. J Exp Bot. 2009. PMID: 19246593 Free PMC article.
-
Thermospermine Synthase (ACL5) and Diamine Oxidase (DAO) Expression Is Needed for Zygotic Embryogenesis and Vascular Development in Scots Pine.Front Plant Sci. 2019 Dec 20;10:1600. doi: 10.3389/fpls.2019.01600. eCollection 2019. Front Plant Sci. 2019. PMID: 31921249 Free PMC article.
-
Moderate stress responses and specific changes in polyamine metabolism characterize Scots pine somatic embryogenesis.Tree Physiol. 2016 Mar;36(3):392-402. doi: 10.1093/treephys/tpv136. Epub 2016 Jan 19. Tree Physiol. 2016. PMID: 26786537 Free PMC article.
-
Involvement of polyamines in the post-anthesis development of inferior and superior spikelets in rice.Planta. 2008 Jun;228(1):137-49. doi: 10.1007/s00425-008-0725-1. Epub 2008 Mar 14. Planta. 2008. PMID: 18340459
References
-
- Acosta C, Pérez-Amador MA, Carbonell J, Granell A (2005) The two ways to produce putrescine in tomato are cell-specific during normal development. Plant Sci 168: 1053–1057
-
- Bagni N, Tassoni A (2001) Biosynthesis, oxidation and conjugation of aliphatic polyamines in higher plants. Amino Acids 20: 301–317 - PubMed
-
- Belmont AS, Braunfeld MB, Sedat JW, Agard DA (1989) Large-scale chromatin structural domains within mitotic and interphase chromosomes in vivo and in vitro. Chromosoma 98: 129–143 - PubMed
-
- Borrell A, Besford RT, Altabella T, Masgrau C, Tiburcio AF (1996) Regulation of arginine decarboxylase by spermine in osmotically-stressed oat leaves. Physiol Plant 98: 105–110 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

